Trajectories of cell-cycle
progression from fixed cell populations
Journal Club 17/02/2017
Luisa Cutillo
Gabriele Gut , Michelle D Tadmor, Dana Pe’er
Lucas Pelkmans & Prisca Liberali
What is this about?
Cycler is a method that constructs a trajectory of cell cycle progression (a Cell Cycle Trajectory or CCT) from fixed images of single cells growing in heterogeneous microenvironments
-
Step 1: Sample Preparation
-
Step 2: Image Processing & Quantification
- Step 3: Build a Cell Cycle Trajectory
Step 1: Sample Preparation
specific protocol to prepare the sample for image acquisition to later construct a Cell Cycle Trajectory with Cycler.
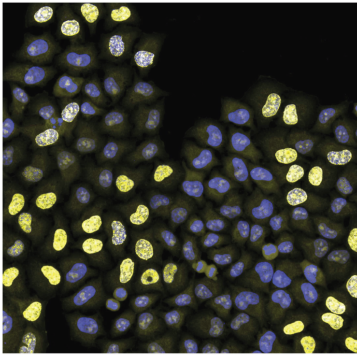
Text
Cell population is stained for:
- DNA (blue signal)
- DNA replication (strong yellow nuclear signal)
- cell outline (weak yellow cytoplasmic signal)
Step 2: Image Processing & Quantification
image analysis protocol in CellProfiler, (which results in a MatLab file) that can be imported into Cycler for classification of G1, S, G2 and M cells, and the construction of a Cell Cycle Trajectory
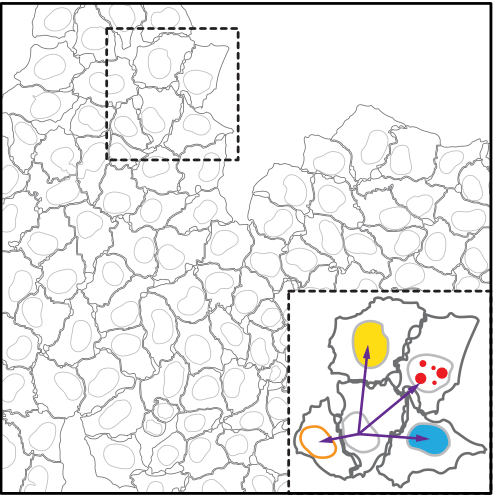
Step 2: Image Processing & Quantification

Nuclei were segmented using images from the DAPI staining
Segmentation of cells and nuclei as well as measurement of single cell features were performed by standard modules of CellProfiler
five features are obtained from CellProfiler modules (MeasureObjectIntensity,MeasureTexture, MeasureObjectAreashape):
nuclei area (corrected for local cell crowding), nuclear DAPI content, nuclear EdU content , nuclear EdU texture feature 5 and 12
Step 2: Image Processing & Quantification
one of the five features : nuclei area (corrected for local cell crowding)
Correction of nuclear area for local cell crowding.
Nuclear area of cells was corrected for local cell crowding in order to use it as a feature in Cycler to construct CCT in population of cells growing in heterogeneous microenvironment, as local cell crowding has an effect on nuclear area
Step 3: Build a Cell Cycle Trajectory
Cycler process on data to construct a Cell Cycle Trajectory.
Classification of discrete cell-cycle phases: G1, S, G2 and M.
a combination of two SVM classifiers is used to identify S phase and M phase cells and a Gaussian mixture model to distinguish between G1 and G2 among the remaining cells.
M-phase cells were excluded before the construction of the CCT, as they link early G1 cells to late G2 cells and decrease robustness in the building of the CCT
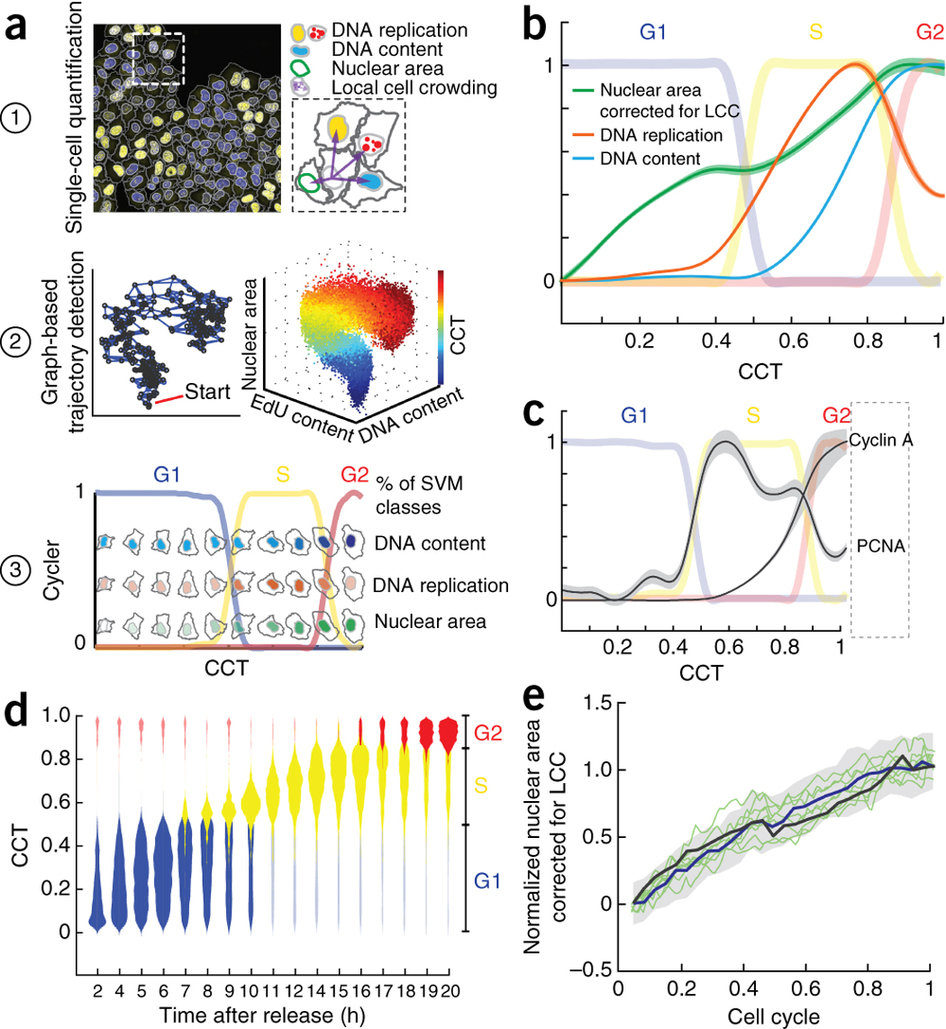
Step 3: Build a Cell Cycle Trajectory
Cycler process on data to construct a Cell Cycle Trajectory.
Cycler is a new version of Wanderlust, performs a k-nearest neighbor graph–based embedding of the multivariate feature space into a single dimension, the cell-cycle trajectory (CCT).
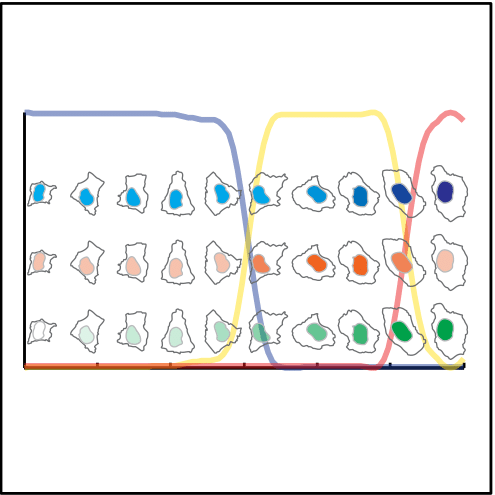
Single cells are ordered to represent the cell cycle. The lines represent the cell cycle phases
G1 S G2
The nuclei of the cells plotted along the CCT show the dynamics of DNA content ( in blue), DNA replication (in red) and nuclear area (in green) along the CCT.
DNA content
DNA replication
Nuclear area
Wanderlust
Wanderlust, a graph-based trajectory detection algorithm that receives multiparameter single-cell events as input and maps them onto a one-dimensional developmental trajectory. Cells are ordered along a trajectory that represents their most likely placement along a developmental continuum.
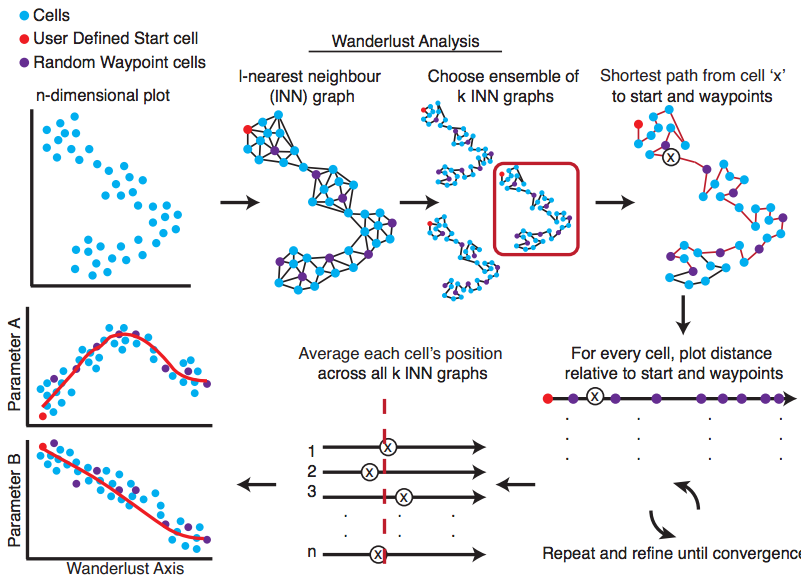
|
Bendall S. et al. Single-Cell Trajectory Detection Uncovers Progression and Regulatory Coordination in Human B cell Development, Cell. 2014. |
nl = 100, dist = euclidean, ng = 5, l = 8, k = 15.
Cycler vs Wanderlust
Cycler has three key changes to the original Wanderlust implementation, focusing on how way-points are used to ensure an accurate trajectory;
-
They found a dense population of cells in the G1 phase, which can account for 60% of the cells. Cycler avoids a uniform selection of waypoints. This would result in a heavy bias of cells at earlier parts of the cell cycle!
-
For each waypoint, its k nearest neighbors are selected and their median computed; the waypoint are then replaced with the cell closest to this median.
- Last, in the formula by which the waypoints refine the placement of all cells along the CCT, Cycler uses a Gaussian-weighting scheme, whereas in Wanderlust, the average is linearly weighted by the distance of the waypoint from the cell.
Robustness of Cycler to the selection of the start population.
Influence of the start-cell parameter on the Cycler algorithm output was tested using Cycler output as a baseline trajectory and re-running Cycler ten times.
Each time, the input start cell was shifted by 0.1 across the baseline trajectory:
As long as the start cell remained in the first 50% of the baseline trajectory, the two trajectories coincide (Pearson's ρ > 0.99)
CCTs for Multiple Cell Lines
Parameters to construct the CCTs were unchanged between HeLa, A431, HEK293, RPEI and COS-7 cells.
The start population for each cell line was gated individually, but each consisted of G1 cells with the smallest nuclear area and the lowest DNA content.
Cycler vs density based methods
Authors show that Cycler outperformed density-based methods in the conditions tested.
Density-based methods cannot account for heterogeneous microenvironments (crowding of cells), whereas Cycler robustly dealt with such images. Note that Cycler may aggregate as many features as desired, thus achieving higher and higher resolution ( used five features).
Density-based methods become prohibitively computationally expensive, as dimensionality increases.