\text{An Investigation of Unsupervised Cell Tracking }
\text{and Interactive Fine-Tuning }
\text{Bio-Image Computing Workshop, ICCV 2025}
\text{HHMI Janelia Research Campus}
\text{Manan Lalit}
\text{Jan Funke}


\text{A typical cell tracking workflow}
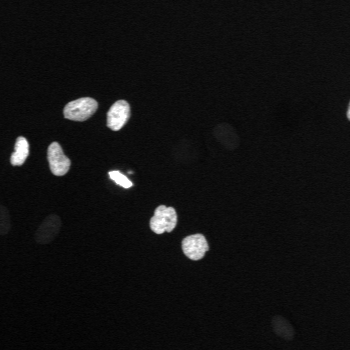
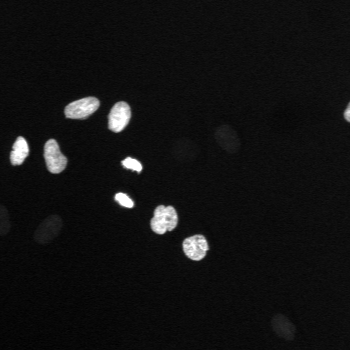
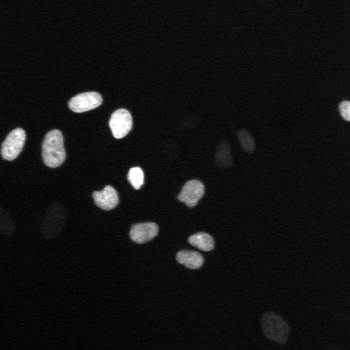
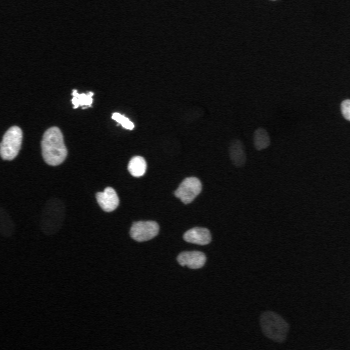
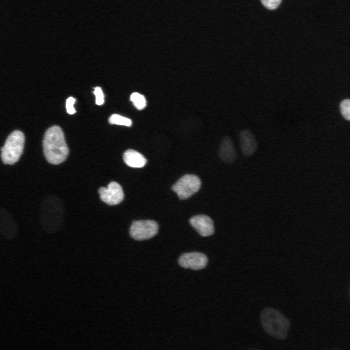
\text{t}
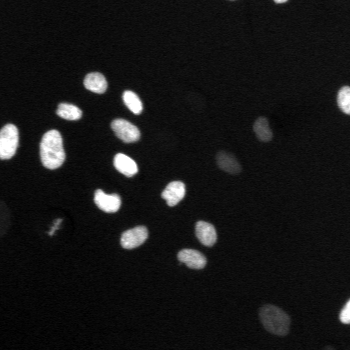
\text{A typical cell tracking workflow}





\text{t}

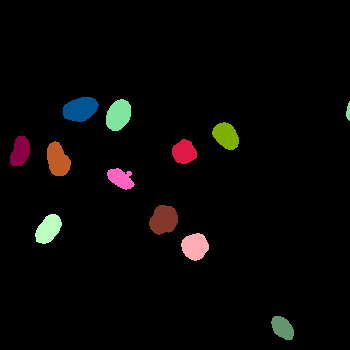
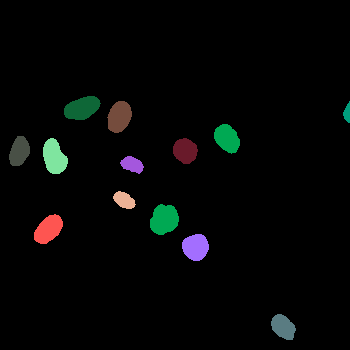

\text{(for e.g. use a pre-trained}
\text{instance segmentation model)}
\text{A typical cell tracking workflow}





\text{t}


\text{(reserve some data for training and testing}
\text{a tracking model)}
\text{Train Subset}
\text{Test Subset}
\text{A typical cell tracking workflow}


\text{t}
\text{(annotate links on the train subset,}
\text{write as an association matrix)}
\text{Train Subset}

























\text{Manually-prepared Association Matrix}
\text{A typical cell tracking workflow}



\text{t}


\text{infer soft-links (association matrices) }
\text{for test subset}

\{

\{
\{

\text{A typical cell tracking workflow}



\text{t}


\text{re-id images}
\text{(for e.g., using a globally constrained }
\text{optimization.)}
\text{A typical cell tracking workflow}





\text{t}

\text{scroll through movie,}
\text{scan for errors and correctly re-id them}.
\text{Instance Segment Cells}
\text{Infer and Re-Id on dataset}
\text{Scroll and Scan for Errors}
\text{Fix Errors}
\text{Prepare GT links for}
\text{training a tracker model}
\text{Incorporate new GT links}
\text{in fine-tuning tracker model}
\text{Instance Segment Cells}
\text{Infer and Re-Id on dataset}
\text{Scroll and Scan for Errors}
\text{Fix Errors}
\text{Prepare GT links for}
\text{training a tracker model}
\text{fine-tune}
\text{2. Strategies which present }
\text{ranked, ambiguous locations}
\text{are needed!}
\text{1. Acquiring link annotations }
\text{is time-consuming!}
\text{3. How do we best incorporate }
\text{new annotations}
\text{for fine-tuning model?}
\text{Instance Segment Cells}
\text{Infer and Re-Id on dataset}
\text{Scroll and Scan for Errors}
\text{Fix Errors}
\text{Prepare GT links for}
\text{training a tracker model}
\text{fine-tune}
\text{1. Acquiring link annotations }
\text{is time-consuming!}
\text{Fully, unsupervised }
\text{\textit{Attrackt} loss.}
\text{2. Strategies which present }
\text{ranked, ambiguous locations}
\text{are needed!}
\text{3. How do we best incorporate }
\text{new annotations}
\text{for fine-tuning model?}
\text{Active Learning Scheme }
\text{which identifies}
\text{cells for proof-reading.}
\text{We present ...}
\text{Fine-tuning Strategy }
\text{which prevents}
\text{over-fitting to few labels.}
\text{Instance Segment Cells}
\text{Infer and Re-Id on dataset}
\text{Scroll and Scan for Errors}
\text{Fix Errors}
\text{Prepare GT links for}
\text{training a tracker model}
\text{fine-tune}
\text{1. Acquiring link annotations }
\text{is time-consuming!}
\text{Fully, unsupervised }
\text{\textit{Attrackt} loss.}
\text{2. Strategies which present }
\text{ranked, ambiguous locations}
\text{are needed!}
\text{3. How do we best incorporate }
\text{new annotations}
\text{for fine-tuning model?}
\text{Active Learning Scheme }
\text{which identifies}
\text{cells for proof-reading.}
\text{We present ...}
\text{Fine-tuning Strategy }
\text{which prevents}
\text{over-fitting to few labels.}
\text{Attrackt Loss}



\text{Step One: Train an auto-encoder with reconstruction loss.}
\text{Attrackt Loss}

\text{(Supervised formulation)}
\text{The goal is to learn a forward function that transforms the cell embeddings at $t$}
\text{to become the corresponding cell embeddings at $t{+}k$.}
\text{If this were done in a supervised manner, i.e. $\mathcal{A}^{GT}$ is available, then the goal is to learn $m_{E}$ and $m_{D}$.}
\text{Attrackt Loss}
\text{(Fully Unsupervised)}
\text{If there are predictable changes in cell appearance over time,}
\text{the model will learn to treat the true predecessor of a cell as the signal,}
\text{and this preference will be reflected in the association matrix.}

\text{consider the other unrelated cells as noise,}
\text{Attrackt Loss}
\text{(Fully Unsupervised)}
\text{Trainable tracker follows \textit{Trackastra} model architecture which maps segmentations in a spatio-temporal}

\text{``Trackastra: Transformer-based cell tracking for live-cell microscopy", Gallusser and Weigert, 2024.}
\text{window to an association matrix.}
\text{Globally constrained optimization}
\text{t}
\text{Build a \textit{candidate graph} by connecting each segmentation at $t$}
\text{to its $k$ nearest spatial neighbors at $t{-}1$.}
\text{Globally constrained optimization}
\text{t}
\text{1. Build a \textit{candidate graph} by connecting each segmentation at $t$}
\text{to its $k$ nearest spatial neighbors at $t{-}1$.}

\text{2. Solve for } \hat{X}_{\mathrm{a}}, \hat{X}_{\mathrm{d}}, \hat{X}_{\mathrm{e}} = \mathrm{arg min}_{X} C_{\mathrm{a}} X_{\mathrm{a}} + C_{\mathrm{d}} X_{\mathrm{d}} + C_{\mathrm{e}} X_{\mathrm{e}},
\text{where }X_{\mathrm{a}}, X_{\mathrm{d}}, X_{\mathrm{e}} \in \{0,1\},
\text{and }C_{\mathrm{e}} = w_{1} \left\|d_{1}\right\|_{2} + w_{2} \left\|d_{2}\right\|_{2} + b.
k=3
\text{Multi-Object Tracker using Integer-Linear Equations}
github.com/funkelab/motile
\text{$C_a$ : Cost of appearance of a new tracklet}
\text{$C_d$ : Cost of disappearance of an existing tracklet}
\text{$C_e$ : Cost of an edge}
\text{``Automated reconstruction of whole-embryo cell lineages}
\text{by learning from sparse annotations", Malin-Mayor \text{et al}, 2023.}
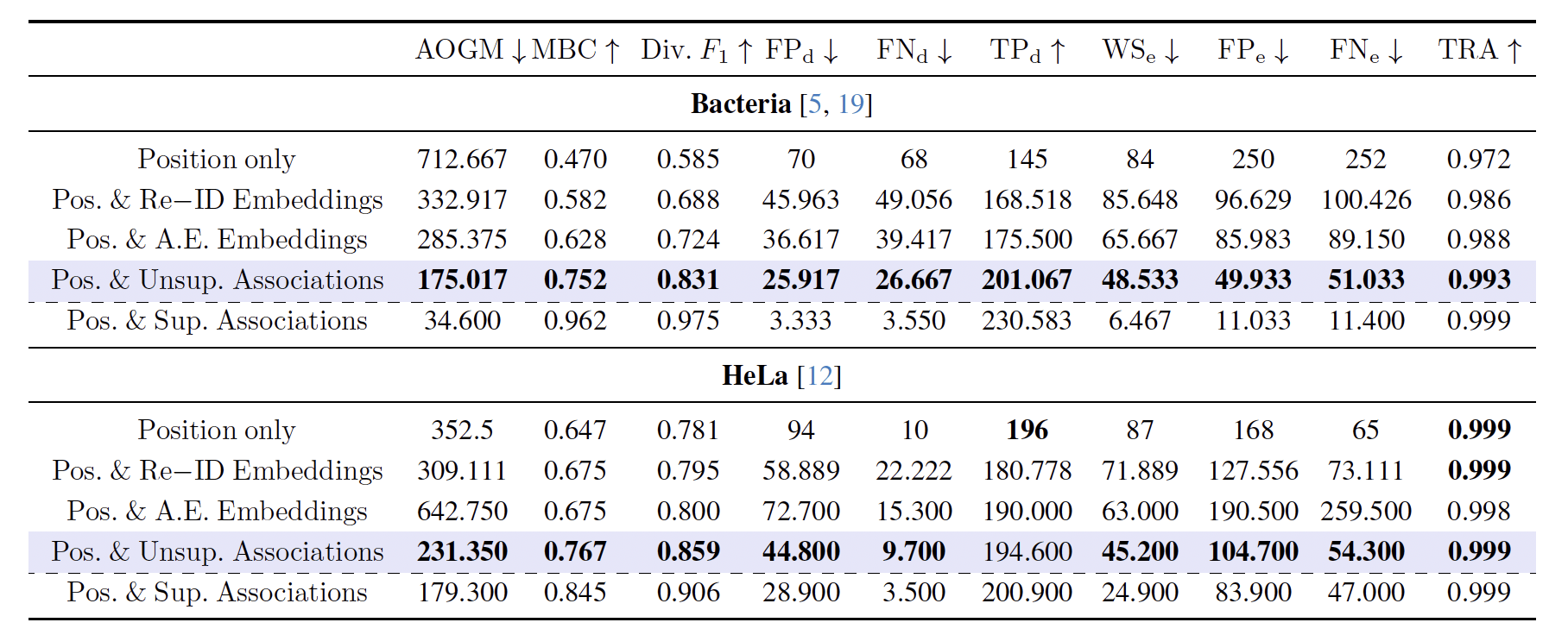
\text{Results: Unsupervised Training}
\text{FP}_{\text{e}}
\text{FN}_{\text{e}}

\text{Results: Unsupervised Training}
\text{Visualizations created using:}
github.com/bentaculum/divisualisation
\text{FP}_{\text{e}}
\text{FN}_{\text{e}}
\text{Instance Segment Cells}
\text{Infer and Re-Id on dataset}
\text{Scroll and Scan for Errors}
\text{Fix Errors}
\text{Prepare GT links for}
\text{training a tracker model}
\text{fine-tune}
\text{1. Acquiring link annotations }
\text{is time-consuming!}
\text{Fully, unsupervised }
\text{\textit{Attrackt} loss.}
\text{2. Strategies which present }
\text{ranked, ambiguous locations}
\text{are needed!}
\text{3. How do we best incorporate }
\text{new annotations}
\text{for fine-tuning model?}
\text{Active Learning Scheme }
\text{which identifies}
\text{cells for proof-reading.}
\text{We present ...}
\text{Fine-tuning Strategy }
\text{which prevents}
\text{over-fitting to few labels.}
\text{Active-Learning Scheme}
\text{Compare against randomly selecting nodes}
\text{score}(v) = \left| \sum \mathcal{A}_{\text{GT}}(v) - \sum \mathcal{A}_{\text{solver}}(v) \right| .
\text{Identify ambiguous nodes! }
\text{Rank nodes based on difference between G.T. and ILP solution. }
\text{Instead use, }\text{score}(v) = \left| \sum \mathcal{A}(v) - \sum \mathcal{A}_{\text{solver}}(v) \right| .
\text{In reality, one doesn't have access to the $\mathcal{A}_{\text{GT}} (v)$. }
\text{(Optimal Sampling)}
\text{(Confidence-based Sampling)}
\text{(Random Sampling)}
\text{$\sum \mathcal{A}_{\text{GT}} (v)$: Number of outgoing edges from node $v$ in G.T.}
\text{$\sum \mathcal{A}_{\text{solver}} (v)$: Number of outgoing edges from node $v$ in ILP solution}
\text{$\sum \mathcal{A} (v)$: Sum of ougoing associations for node $v$ predicted by}
\text{unsupervised tracker.}
\text{Instance Segment Cells}
\text{Infer and Re-Id on dataset}
\text{Scroll and Scan for Errors}
\text{Fix Errors}
\text{Prepare GT links for}
\text{training a tracker model}
\text{fine-tune}
\text{1. Acquiring link annotations }
\text{is time-consuming!}
\text{Fully, unsupervised }
\text{\textit{Attrackt} loss.}
\text{2. Strategies which present }
\text{ranked, ambiguous locations}
\text{are needed!}
\text{3. How do we best incorporate }
\text{new annotations}
\text{for fine-tuning model?}
\text{Active Learning Scheme }
\text{which identifies}
\text{cells for proof-reading.}
\text{We present ...}
\text{Fine-tuning Strategy }
\text{which prevents}
\text{over-fitting to few labels.}
\text{Interactive Fine-Tuning}
\text{1. Use LoRA to train a fraction of weights.}
\text{2. Fine-tuning objective: $\mathcal{L}_{\text{fine-tuning}} = \mathcal{L}_{\text{unsup.}} + \mathcal{L}_{\text{sup.}} + \mathcal{L}_{\text{pseudo-sup.}}$}
\text{``LoRA: Low-Rank Adaptation of Large Language Models", Hu \textit{et al}, 2021.}
\text{Compare predicted associations for G.T. associations}
\text{available for top $k$ ambiguous nodes.}
\text{Simulate proof-reading by selecting the top $k$ ambiguous nodes }
\text{and using the outgoing G.T. links for these nodes. }
\text{Interactive Fine-Tuning}
\text{1. Use LoRA to train a fraction of weights.}
\text{2. Fine-tuning objective: $\mathcal{L}_{\text{fine-tuning}} = \mathcal{L}_{\text{unsup.}} + \mathcal{L}_{\text{sup.}} + \mathcal{L}_{\text{pseudo-sup.}}$}
\text{``LoRA: Low-Rank Adaptation of Large Language Models", Hu \textit{et al}, 2021.}
\text{Compare predicted associations with associations}
\text{inferred by solver and available for bottom $10\%$}
\text{ambiguous nodes.}
\text{Results: Interactive Fine-Tuning}
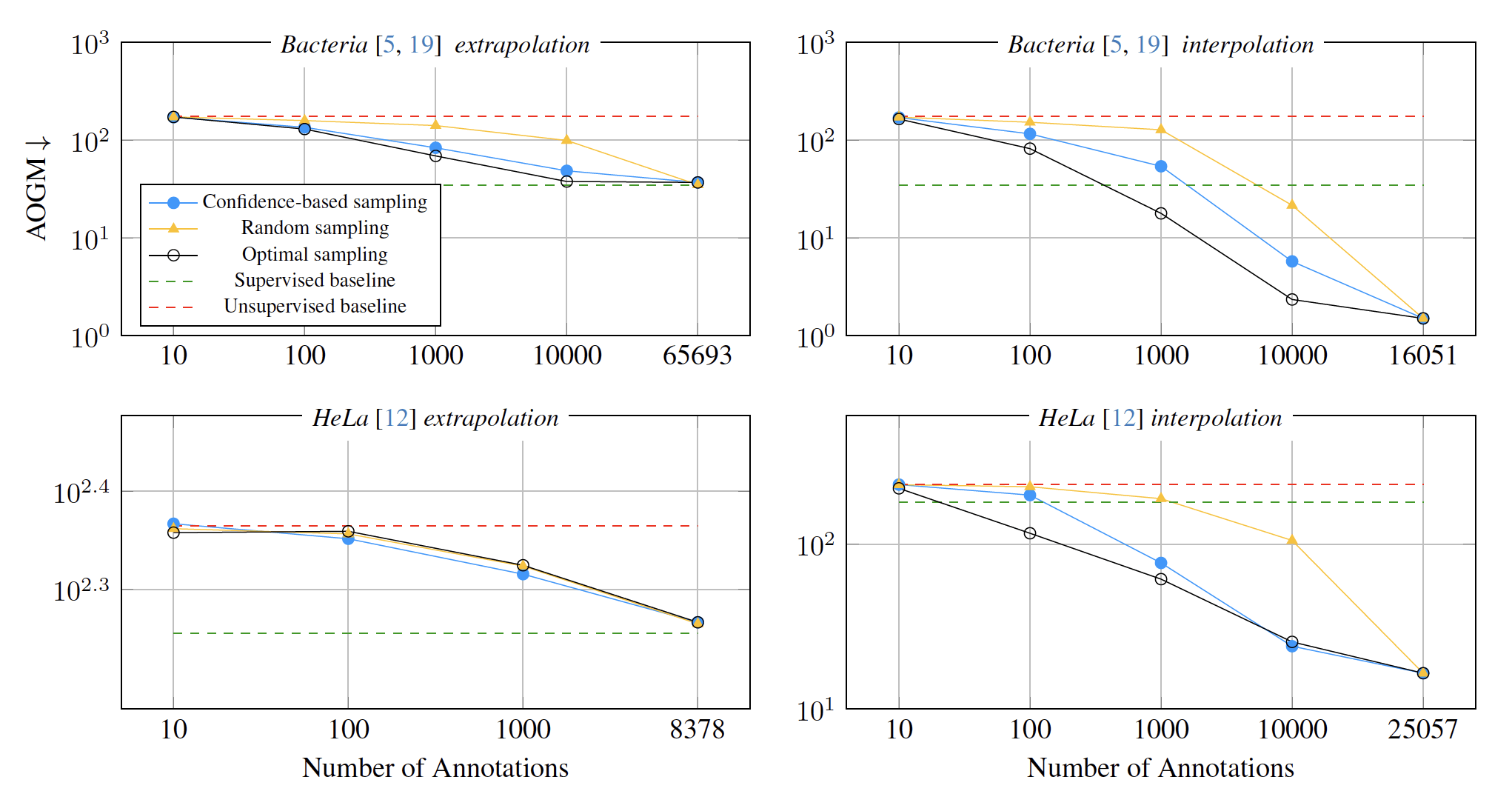
\text{Results: Interactive Fine-Tuning}

\text{Results: Interactive Fine-Tuning}

\text{Results: Interactive Fine-Tuning}

\text{Results: Interactive Fine-Tuning}

\text{Confidence-based sampling outperforms Random sampling.}
\text{Optimal sampling leads to faster AOGM reduction.}
\text{Results: Unsupervised Pre-Training and Fine-Tuning vs Training from Scratch}
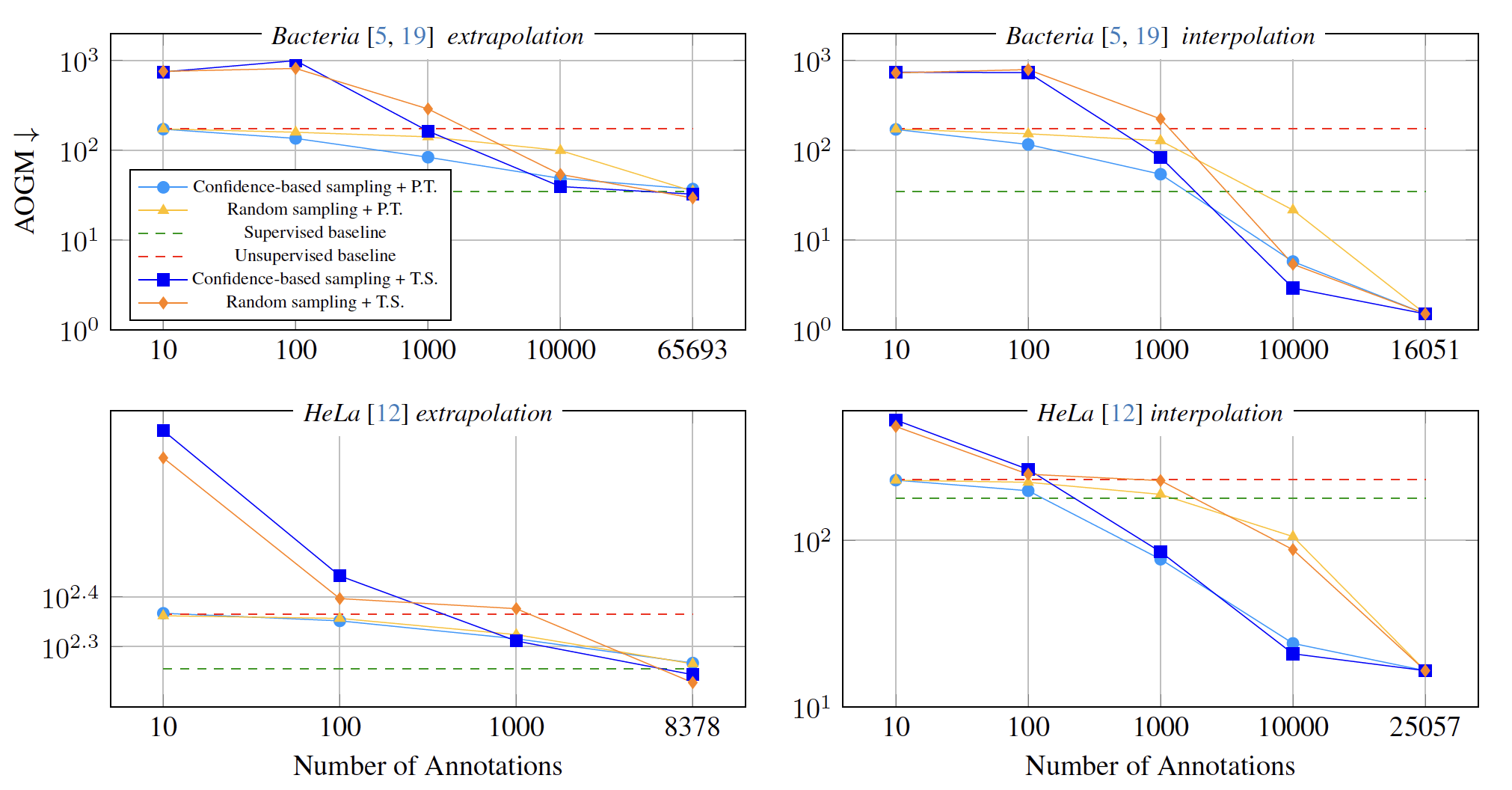
\text{In the low annotation regime, unsupervised pre-training provides a clear advantage.}
\text{Acknowledgements}
\text{Attrackt Code:}
\text{github.com/funkelab/attrackt}
\text{Attrackt Experiments:}
\text{github.com/funkelab/attrackt\_experiments}





\text{Diane Adjavon}
\text{Ben Gallusser}
\text{Jan Funke}
\text{Caroline Malin-Mayor}
\text{Morgan Schwartz}
\text{Check out our \textit{napari} plugin:}
\text{github.com/funkelab/motile\_tracker}