Fluctuations in biofilm topographies

Pablo Bravo
YunkerLab, Georgia Institute of Technology
APS March Meeting 2023
smooth
Extracellular matrix formed of polysaccharides, DNA, and proteins



Surface
Interface
Cells
Biofilms are 3D structures
complex surface-attached


Surface
Interface
Cells

Can we learn about the biofilm development/composition by looking at its topography?
Biofilms are 3D structures
complex surface-attached
What is ?




Profiles are flat. A few cells in amplitude, over thousands of micrometers!

\(500 \mu m\)
biofilm topography
Using white-light interferometry, we can capture the profiles of a growing colonies for extended periods of time
are different between strains
Staphylococcus aureus
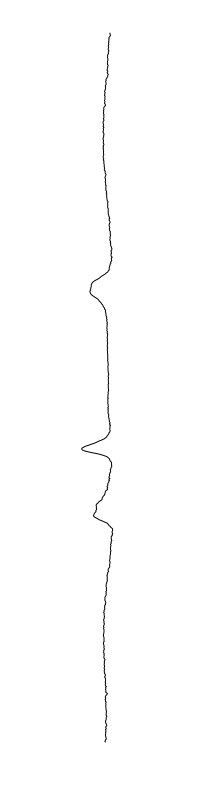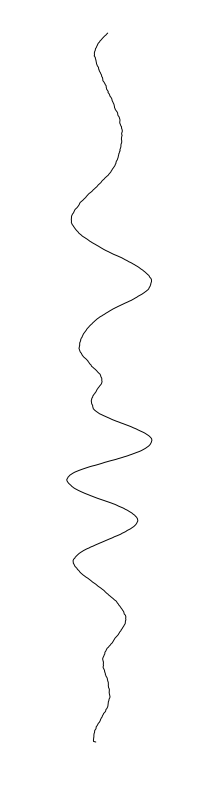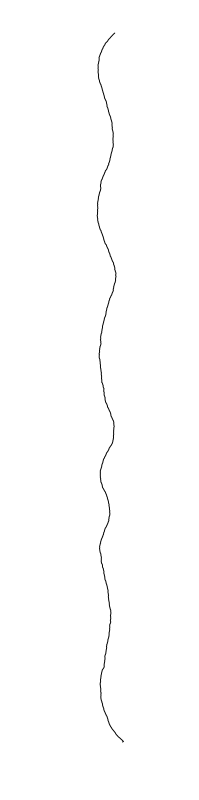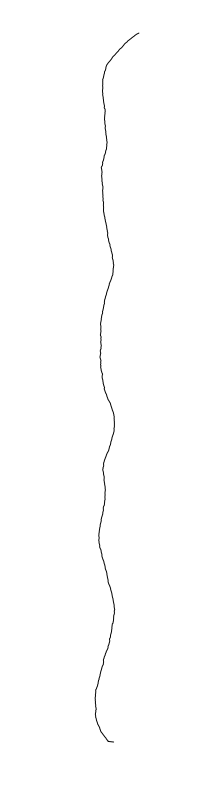
Bacillus cereus
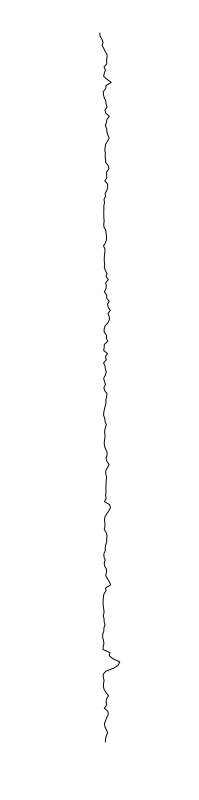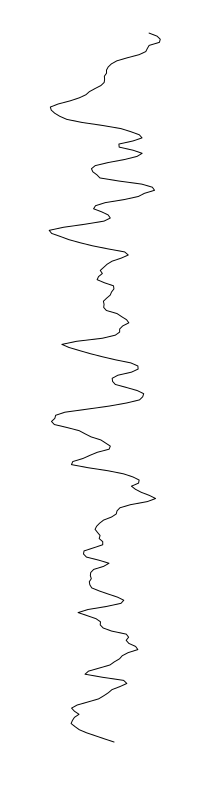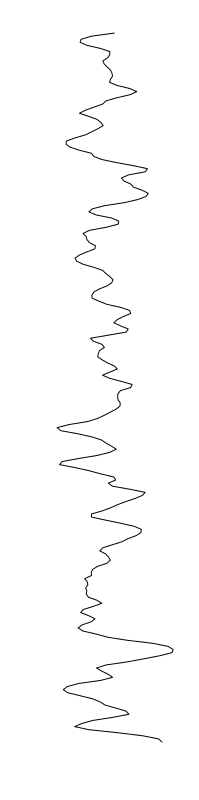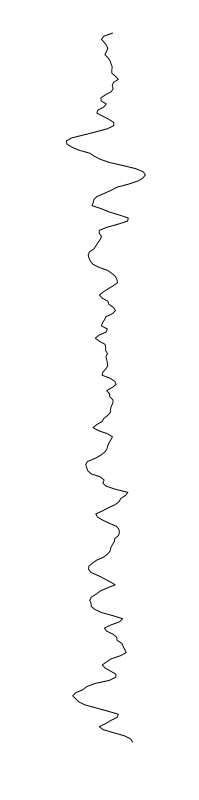
Eschericia coli
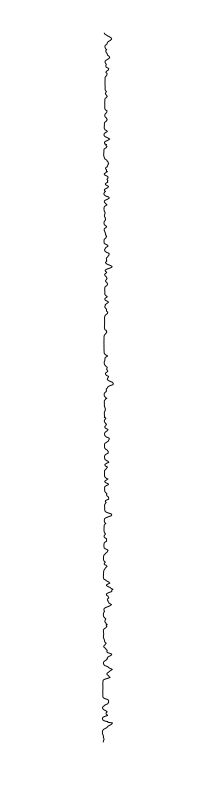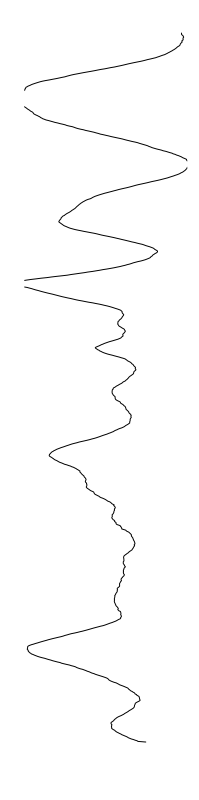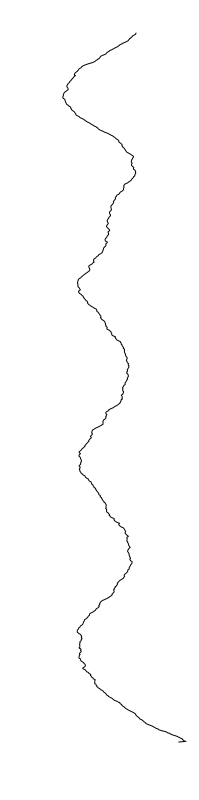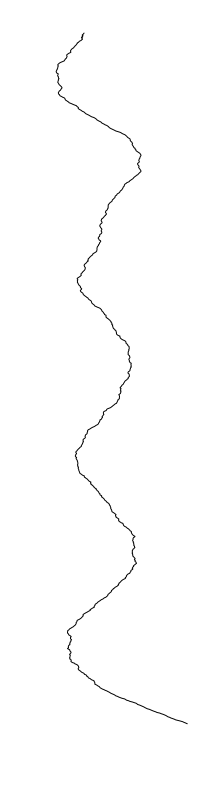
- What is leading to the differences between profiles?
- How do we characterize the dynamics?
\(2 mm\)
\(8 \mu m\)
Time since inoculation [hours]
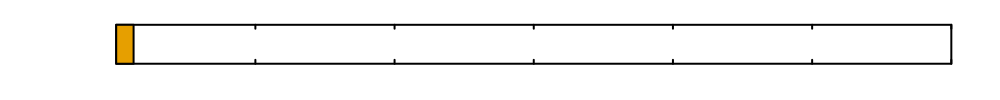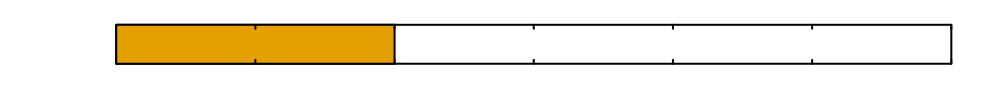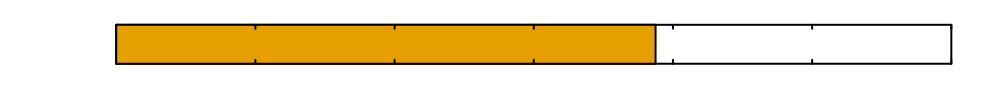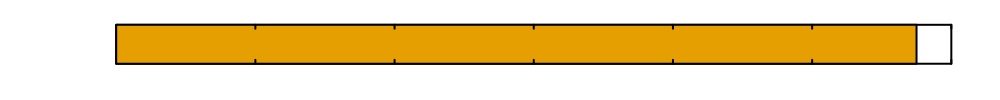
\(0\)
\(24\)
\(48\)
Fluctuation dynamics
Fluctuations can





Aeromonas veronii





Eschericia coli
or
relax
increase
- Width of perpendicular fluctuations in the height profile:
- With a roughness/Hurst scaling exponent \(H\):
- The width saturates at \(l_\text{sat}\), and across the whole sample the width is \(w_\text{sat}\)
\(w_l(t) \propto l^{H} \)
fluctuations
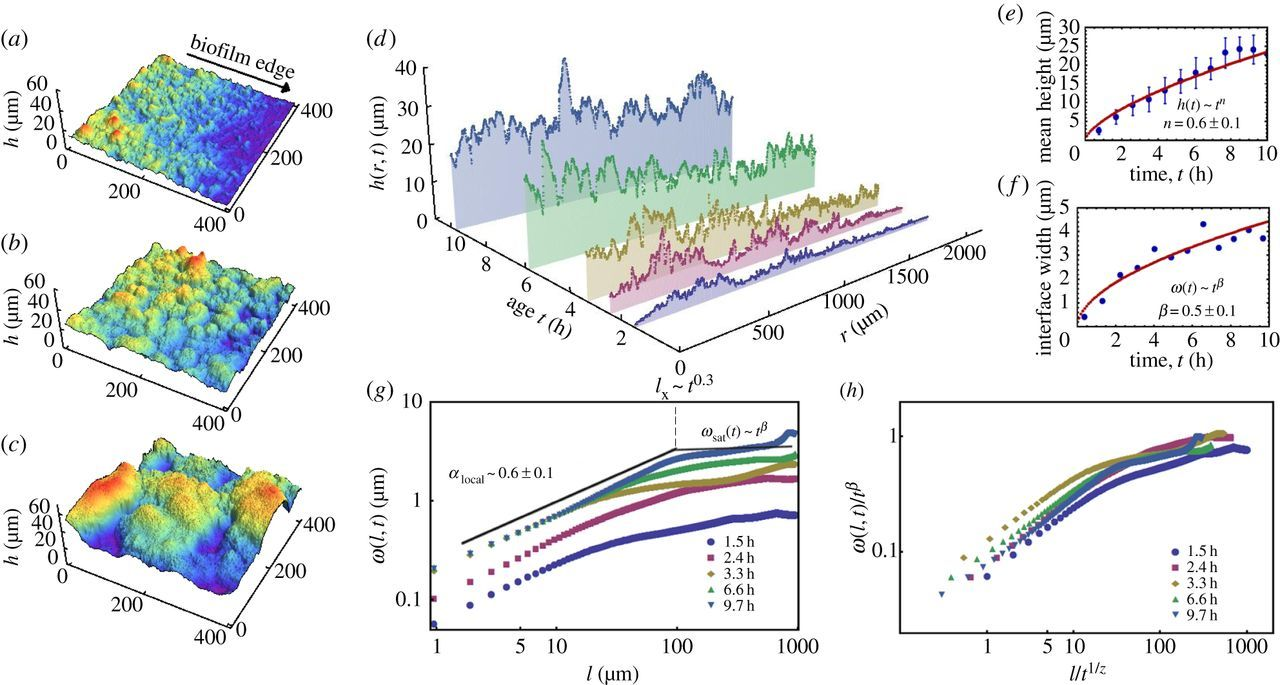
Dervaux, et al. 2014
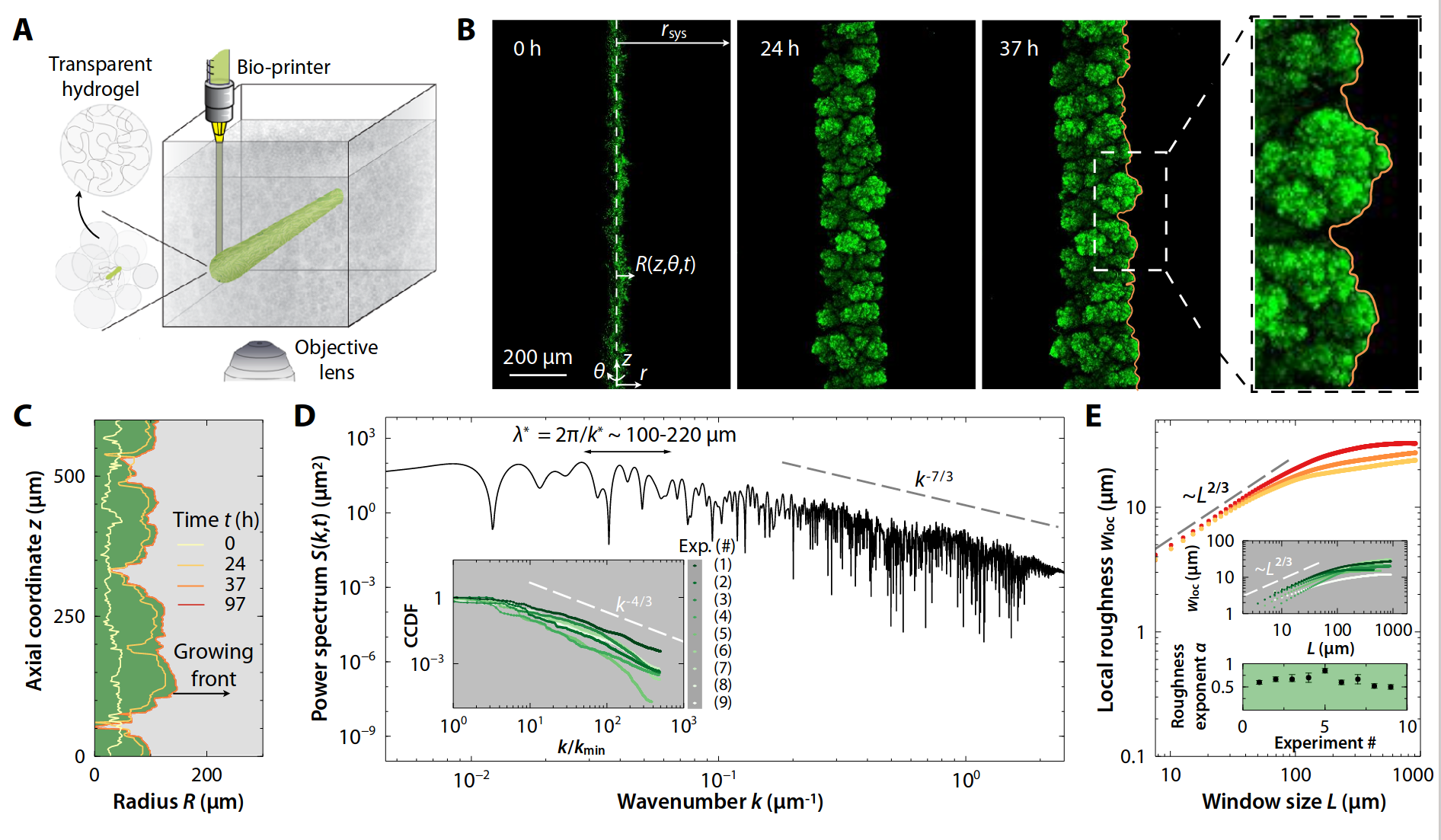
Martinez-Calvo and Bhattacharjee, et al. 2022

\(H\)
\(l_{\text{sat}}\)
\(w_{\text{sat}}\)
Quantifying
across strains


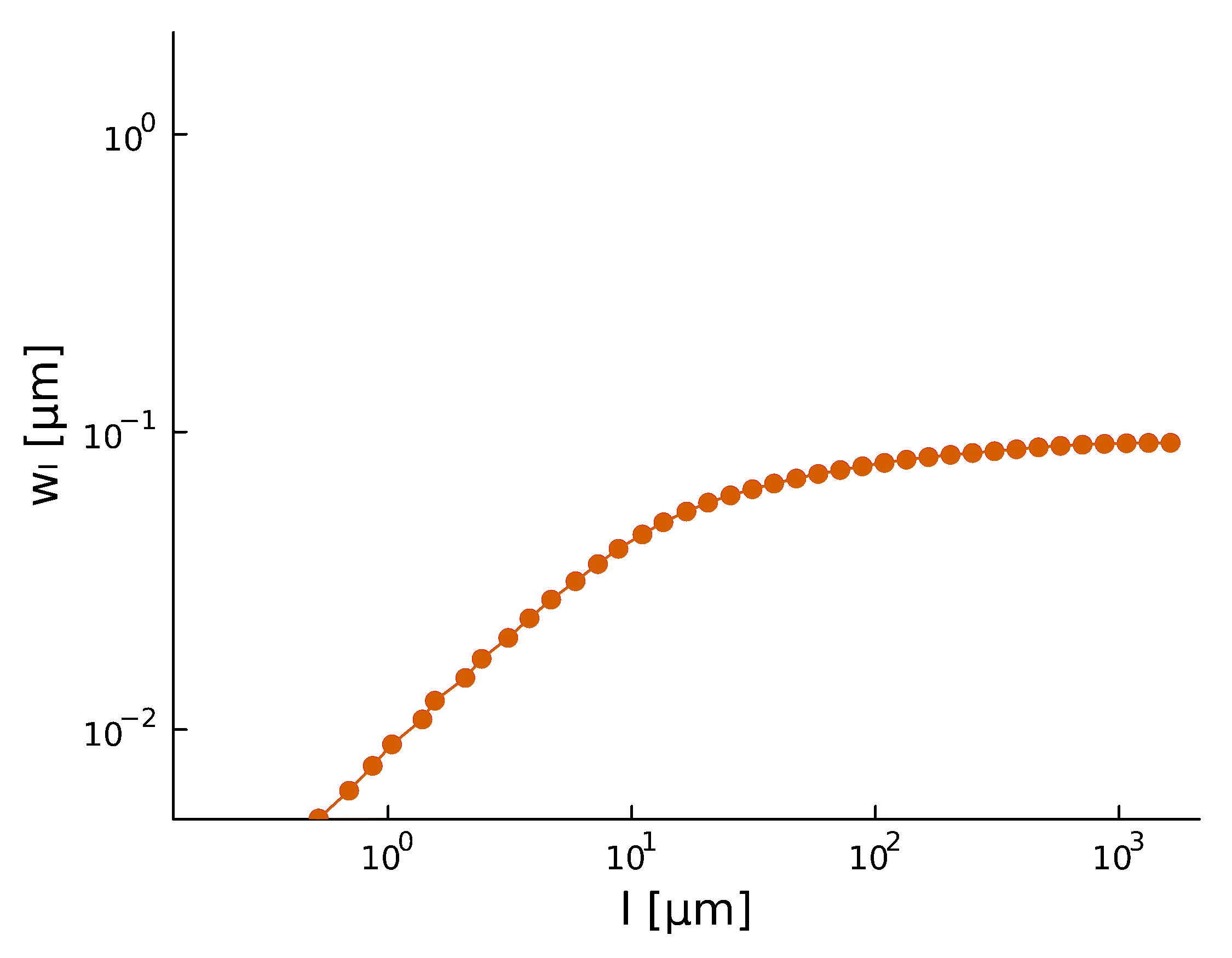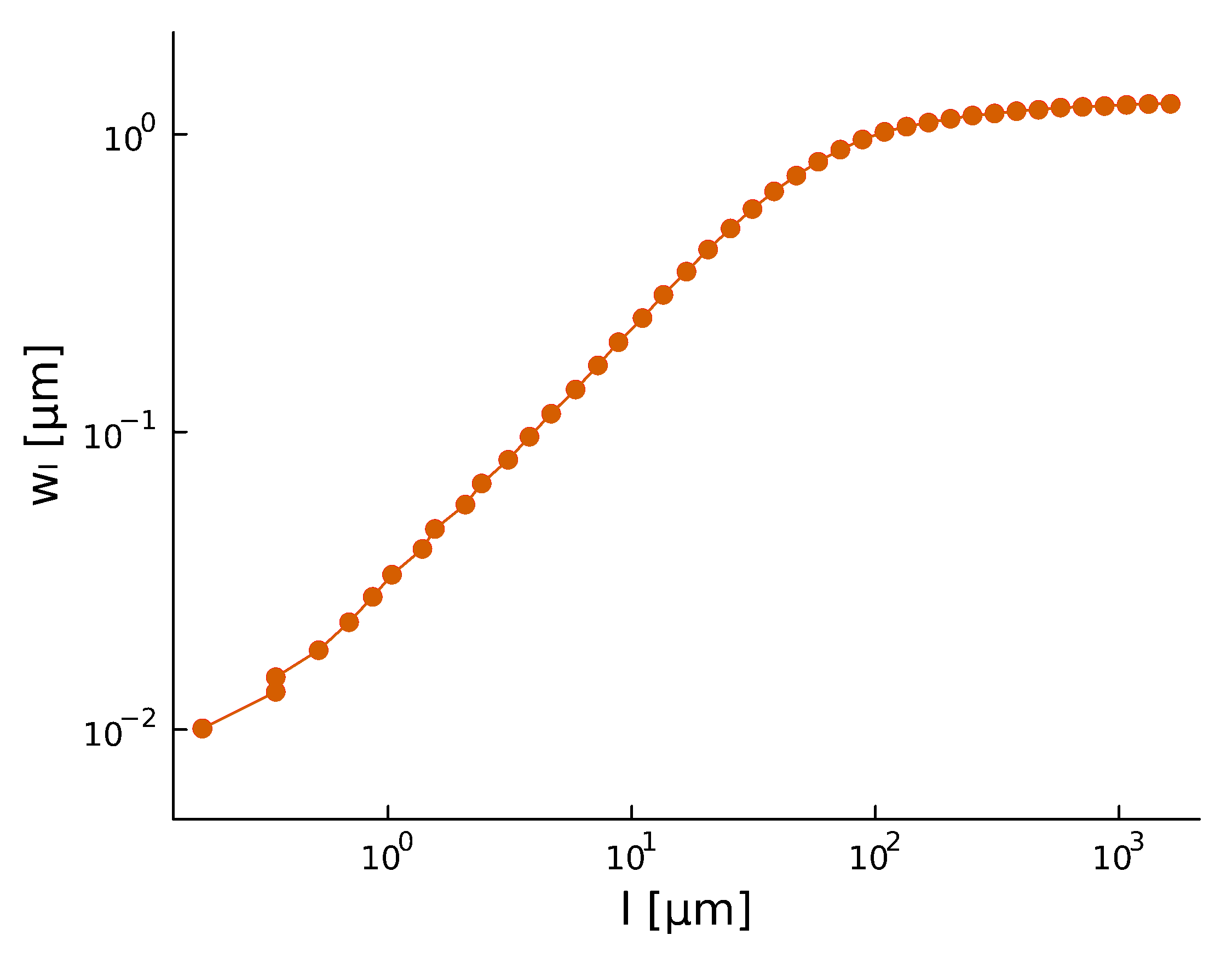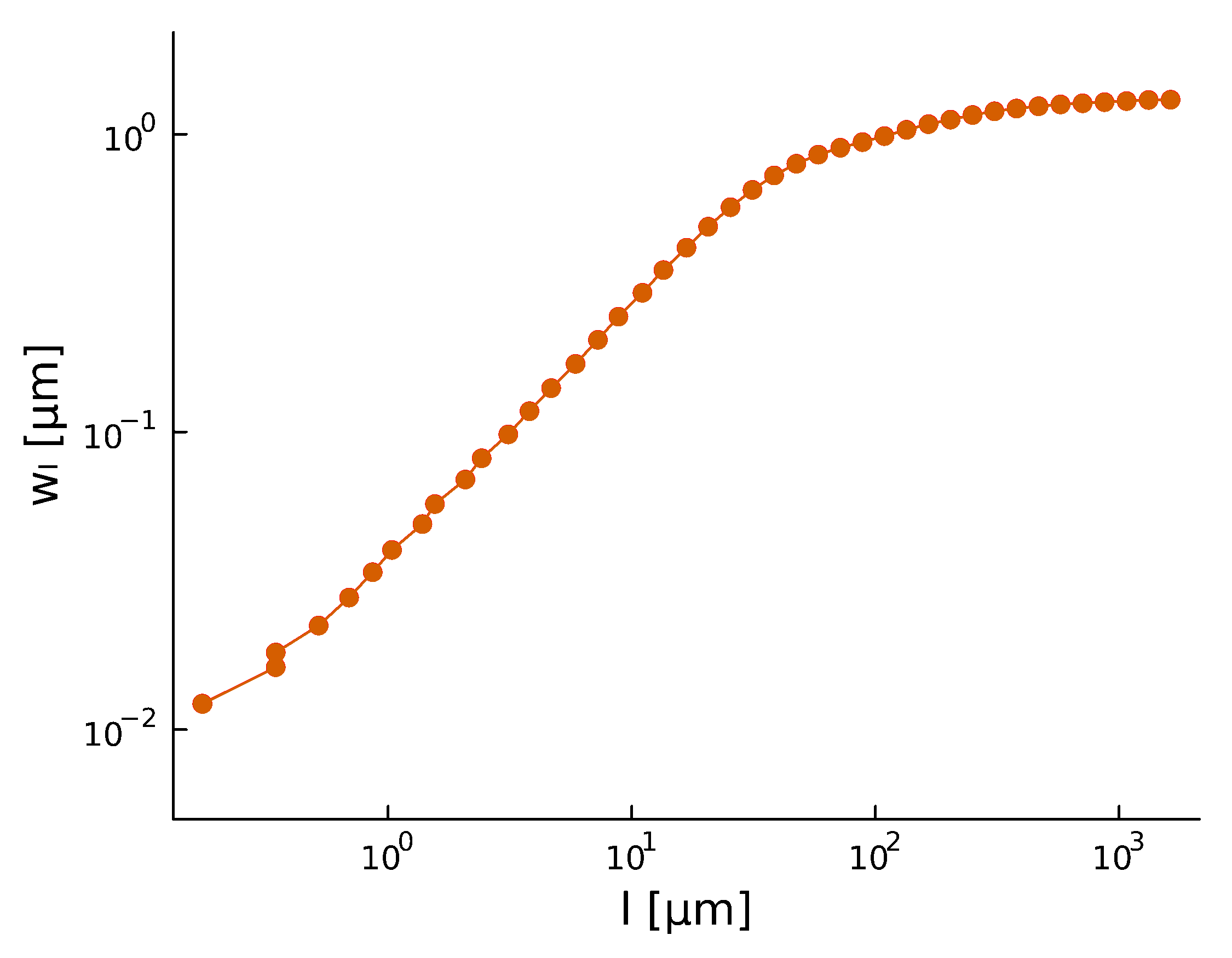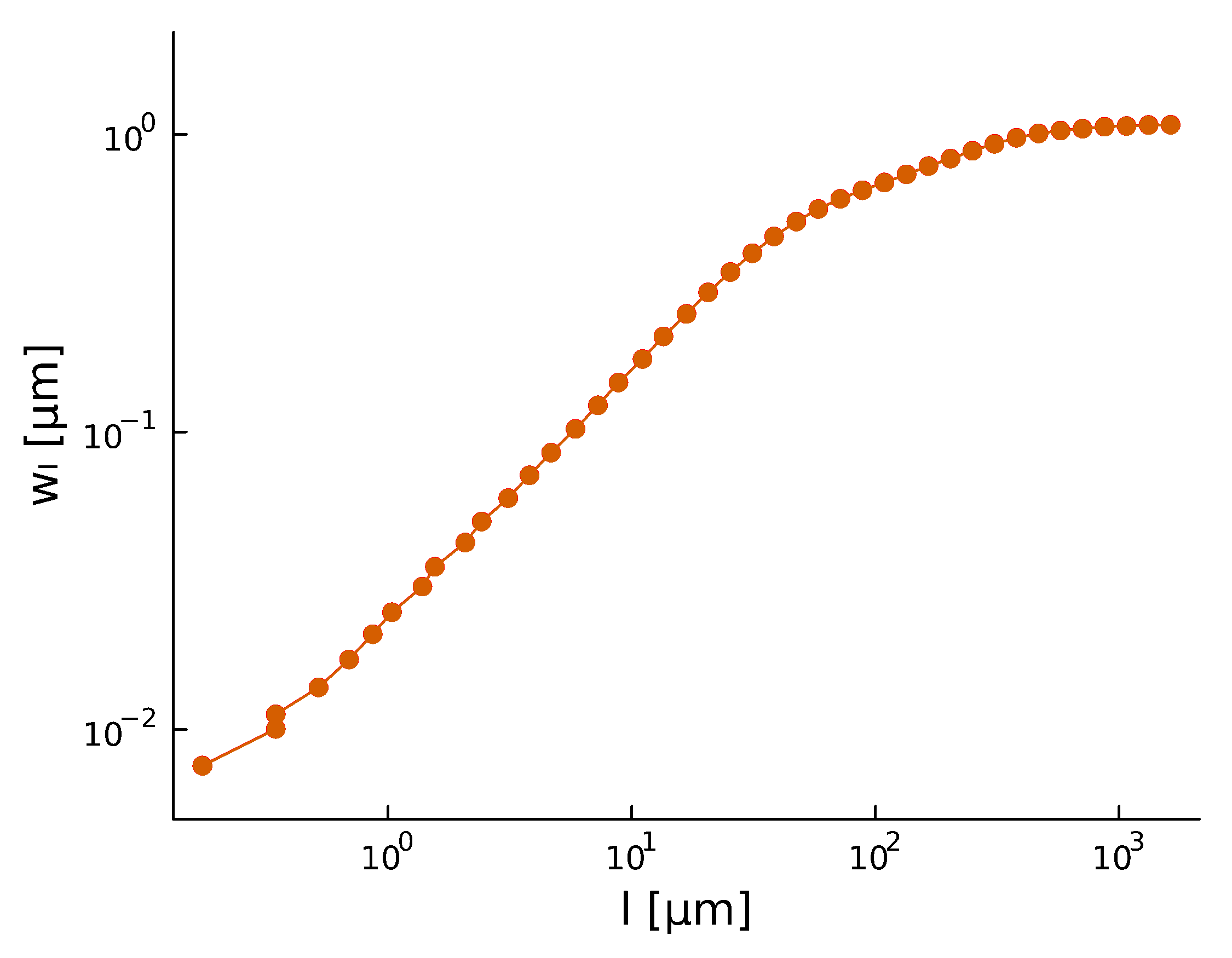
Roughness \(H\), after a period of time, stabilizes at \(H_{\text{steady}} \sim 0.8\)
Roughening



and
There is high variability between microbes
\(w_{sat}\)
vertical growth
during biofilm growth



Growth rate
Decay rate
Diffusion length

Two regimes
In press at PNAS




There is high variability between microbes
And an apparent correlation between vertical growth dynamics and the topography!

Colonies reach nutrient depletion length \(L\)
Knowing the moment when the colony is growing the fastest, just by looking at
fluctuations
and
\(w_{sat}\)
vertical growth
Are fluctuations ?

\(S(k) [\mu m^4]\)
\(k [\mu m^{-1}]\)




\(10^4\)
\(10^3\)
\(10^2\)
\(10^1\)
Dynamic scaling, a test for self-affinity requires:
\(\nu = 1+2H\)
We do not see observe dynamic scaling.
It is close but not the same!
scale-free
Testing again
self-affinity



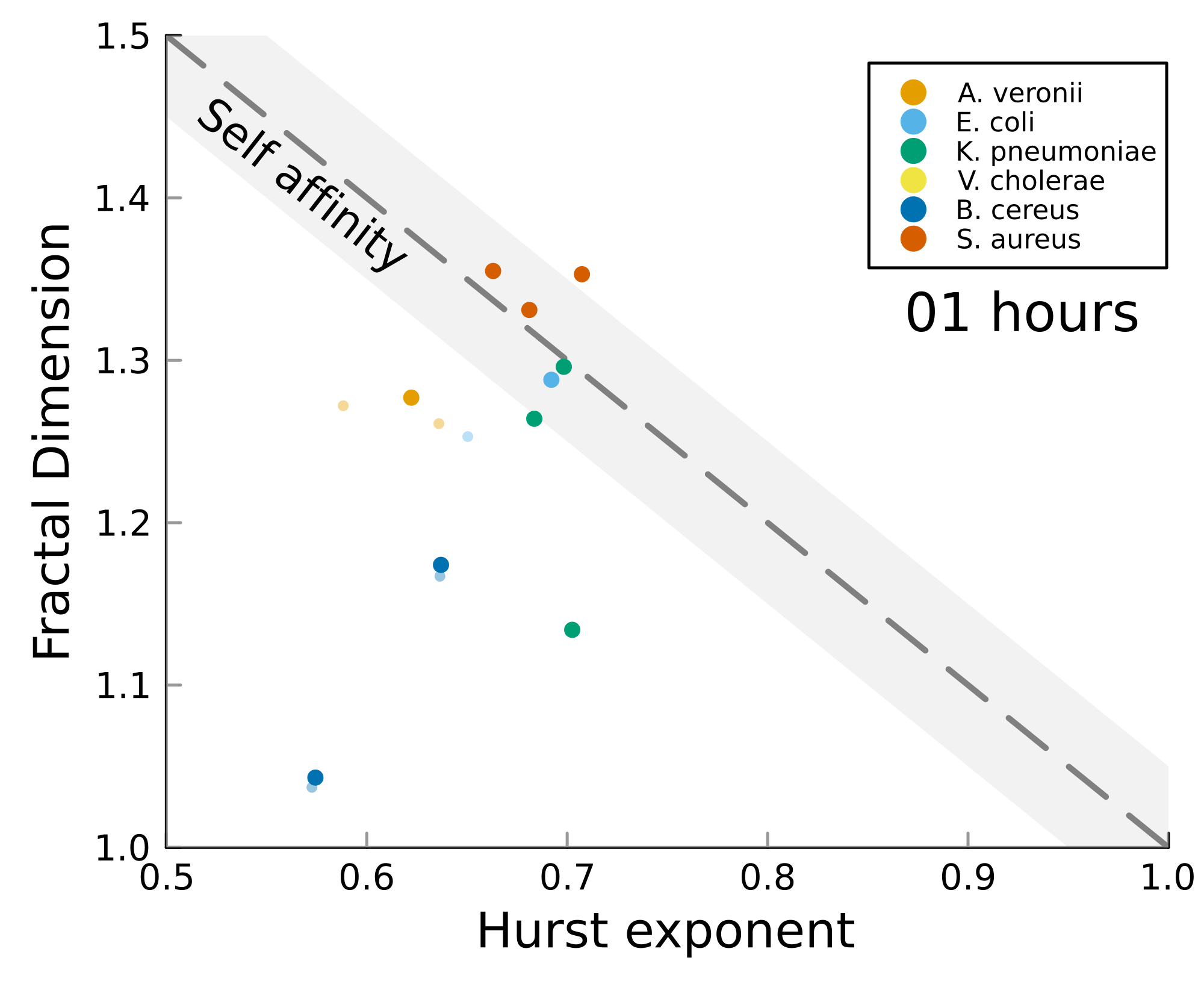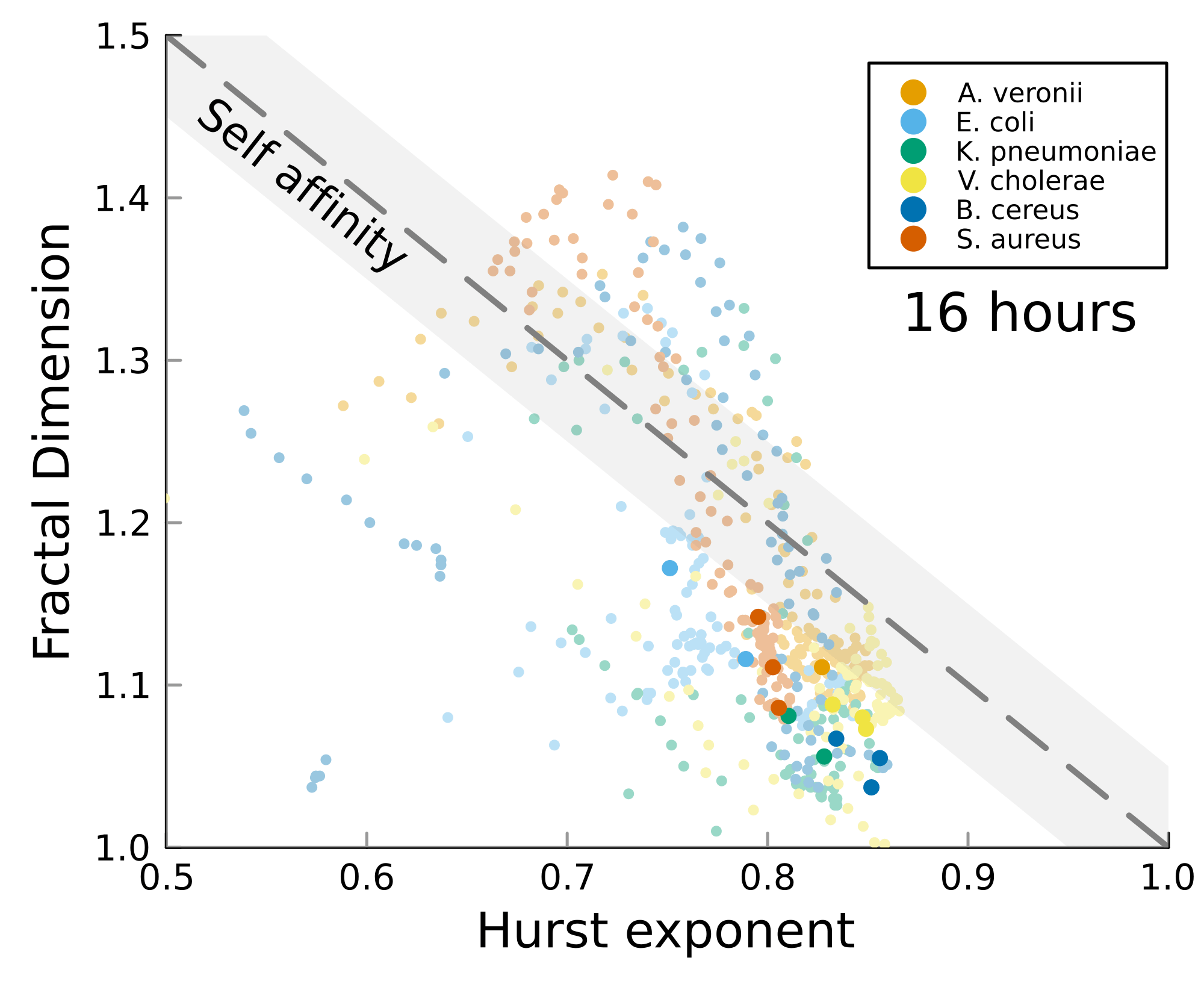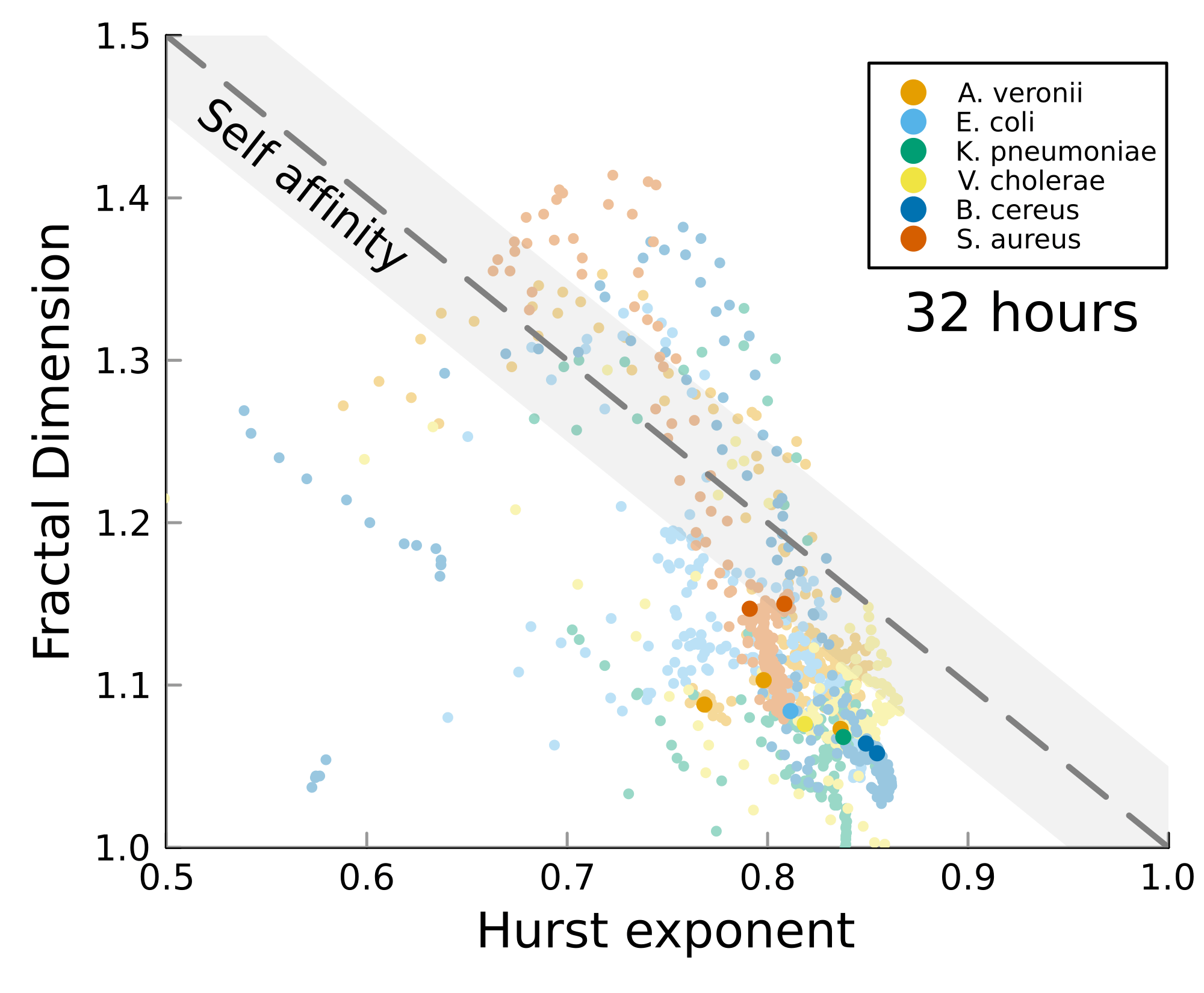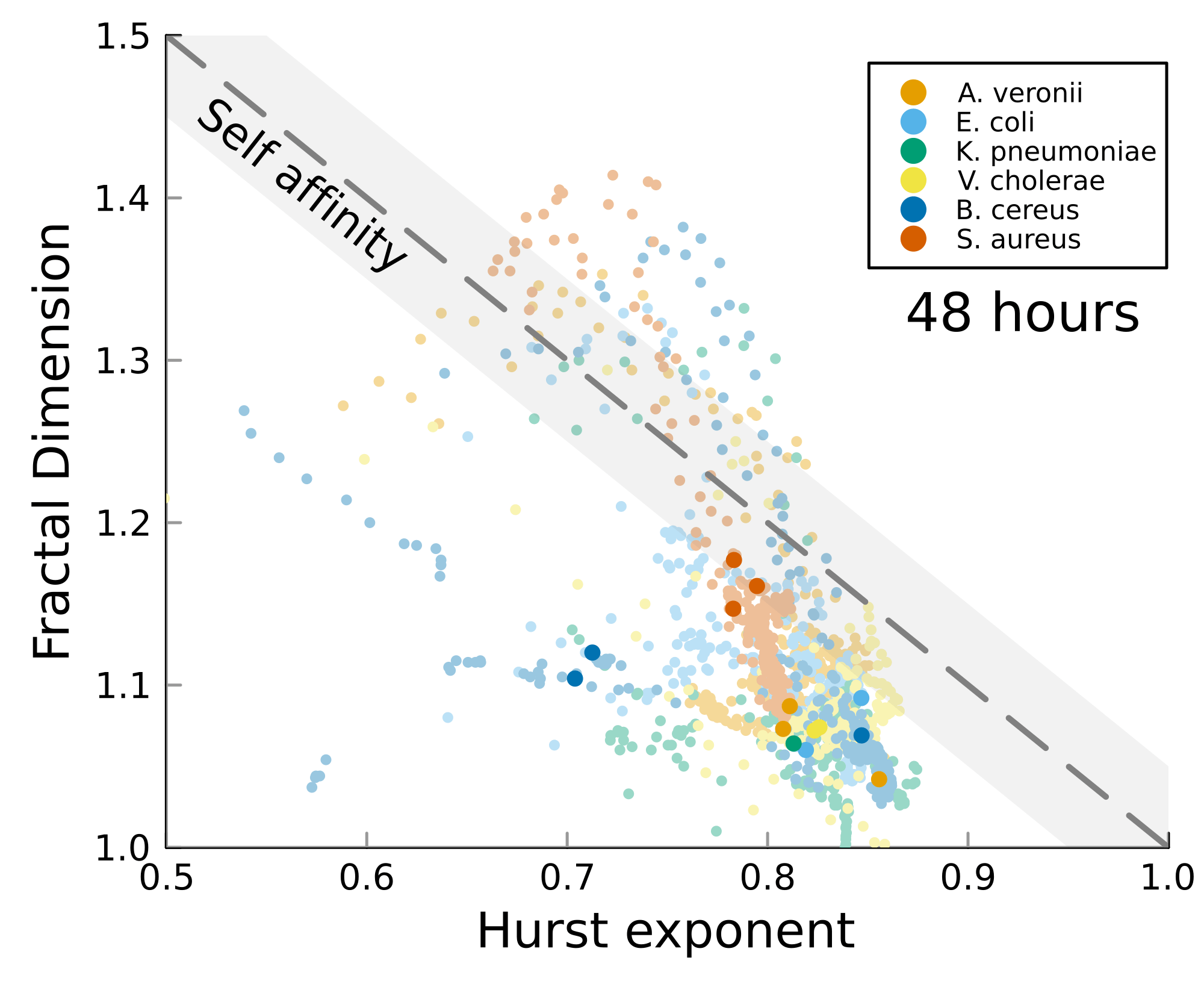
We can test self-affinity using the fractal dimension \(D\)
\(D + H = 2\)
\(D + H = n +1\), where \(n\) is the base dimension of the system
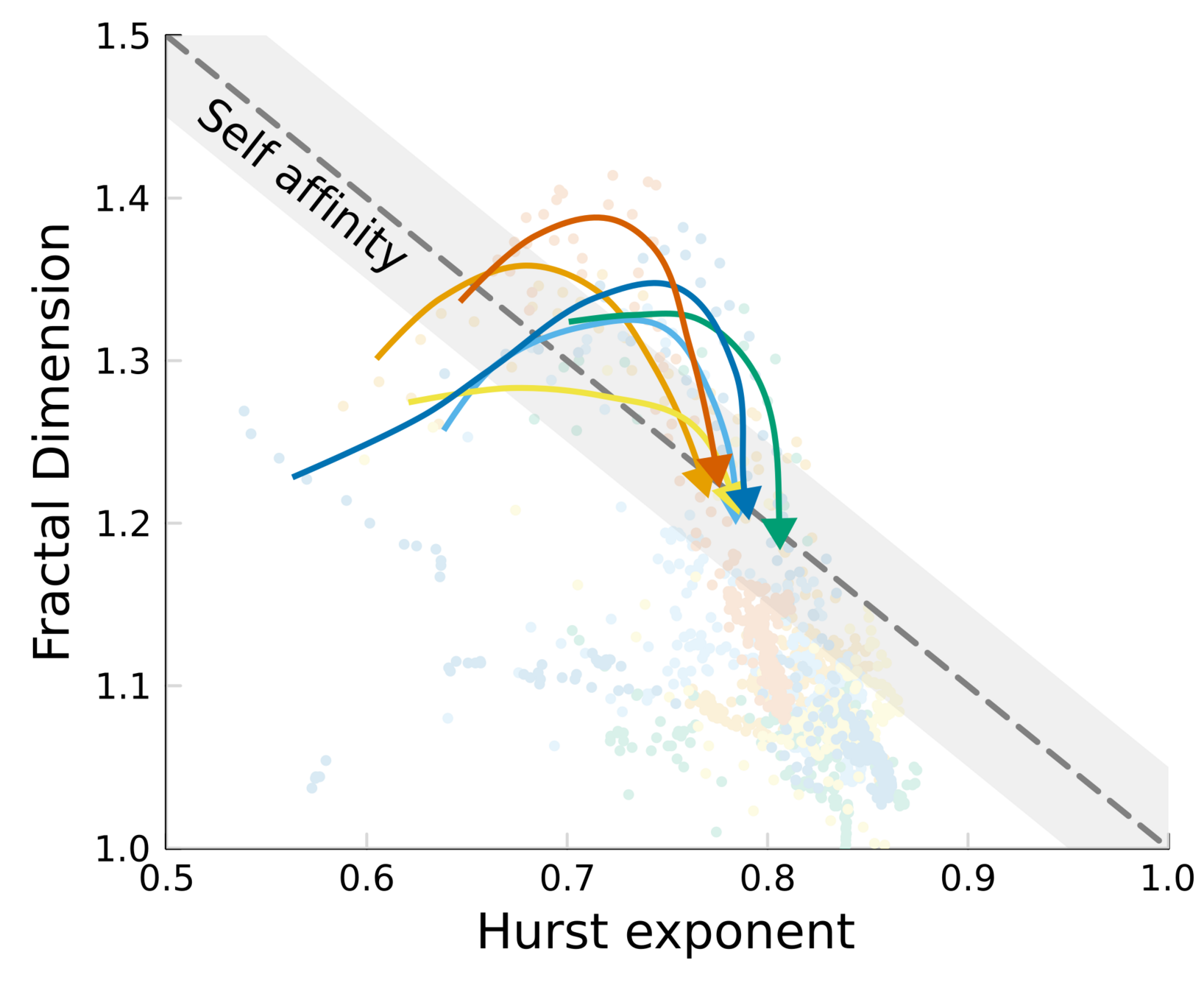


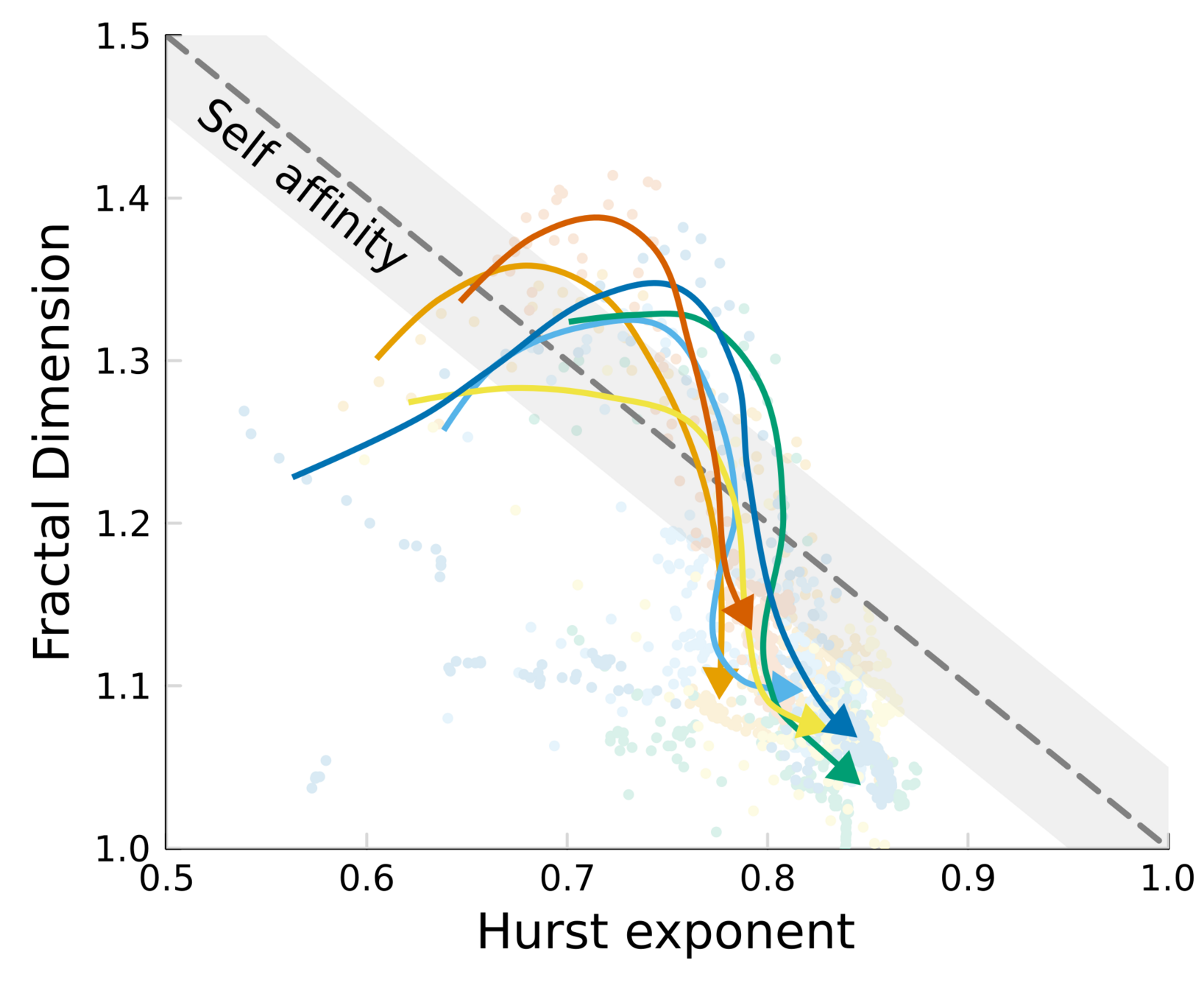
Topography dynamics as a consequence of growth through a viscoelastic material:
- Reach SA while growth is fast
- Relax when the colony is tall, and fluctuations get damped

Acknowledgements


NIH-NIMS
NSF BMAT
Biolocity
Dr. Peter Yunker
Dr. Brian Hammer
Dr. Siu Ling Ng
Dr. Thomas Day
Aawaz Pokhrel
Emma Bingham
Funding
Thanks!
Adam Krueger
Raymond Copeland
Maryam Hejri
Lin Zhao
Chris Zhang


Can we establish a link between and ?
Hopefully!
topographies
biofilm development