Viral phylodynamics of
host adaptation
& antigenic evolution
Sidney Bell
Bedford Lab
Molecular & Cellular Biology





car

cat
Reconstructing evolution is a lot like "telephone"
zebra



....
car
cat
Reconstructing evolution is a lot like "telephone"
zebra




Reconstructing evolution is a lot like "telephone"






ACG
AGG
AGT
Evolution provides context
to viral outbreaks
Where did this virus come from?
When did interesting traits change?




ACG
AGG
AGT
Contextualizing the HIV pandemic
Understanding dengue antigenic evolution
Outline


Contextualizing the HIV pandemic
Cross-species transmission can be devastating



Ebola
Swine flu

HIV
HIV has entered human populations 16 times




x2
x2
x12
45+ primate species carry a unique SIV



?
?
?

How often do
SIVs switch
host species?
Viral phylogenies show
history and migration
Color = host species
Bell & Bedford, PLoS Pathog., 2017
0.5 sub/site
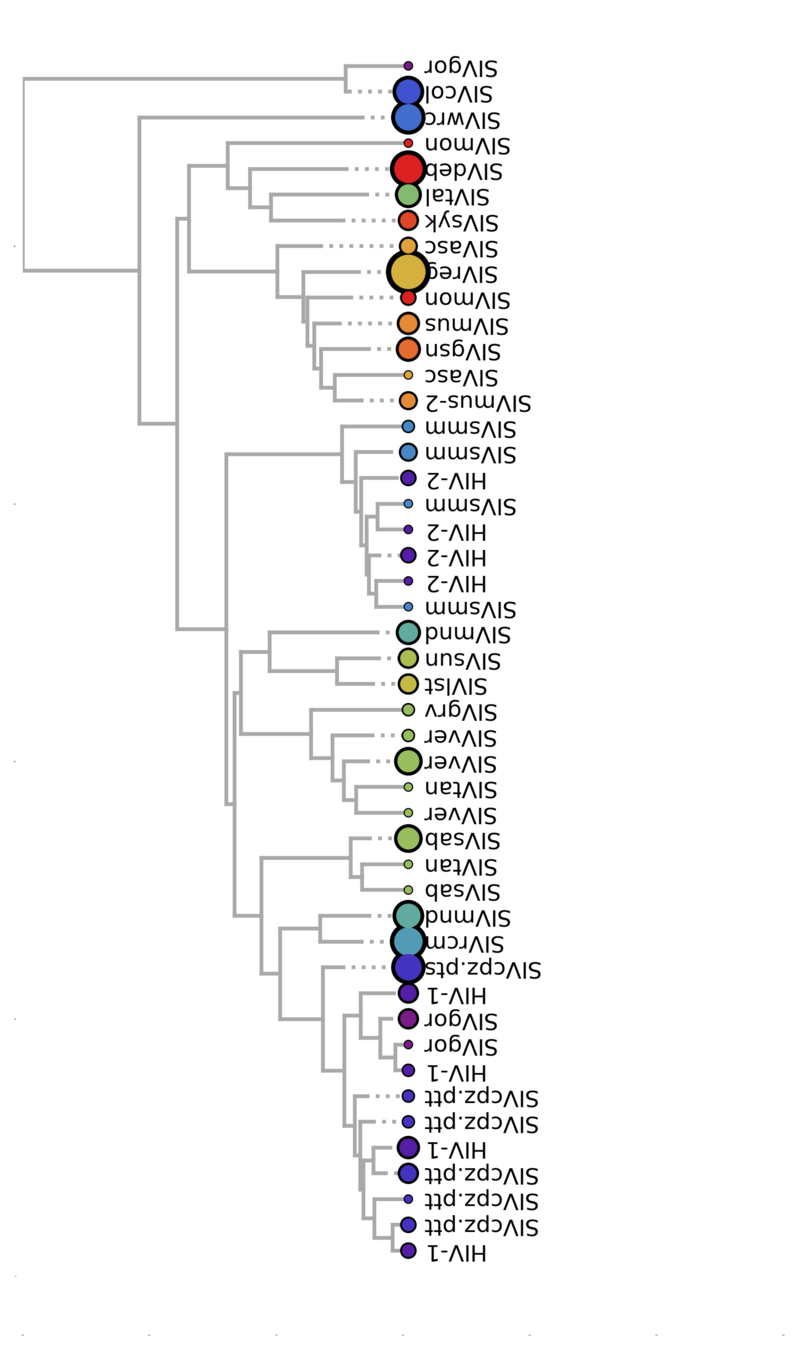

Cross-species transmission looks
like color changes
Caveat: recombination complicates evolutionary history
+
Recombination


Solution: assess each genome segment separately



12 segments x ~2,000 trees = 24,000 trees
Assessing many hypotheses
Bell & Bedford, PLoS Pathog., 2017

Modeling host species as a discrete trait
From host
To host
A B C
...
A
B
C
...
SIVs have been adapting to new hosts for millions of years
Bell & Bedford, PLoS Pathog., 2017



Understanding dengue antigenic evolution
Dengue is a mosquito-borne virus
that kills 15,000 children annually

Dengue has recently expanded geographically
There are four serotypes of dengue
DENV1
DENV3
DENV4
DENV2
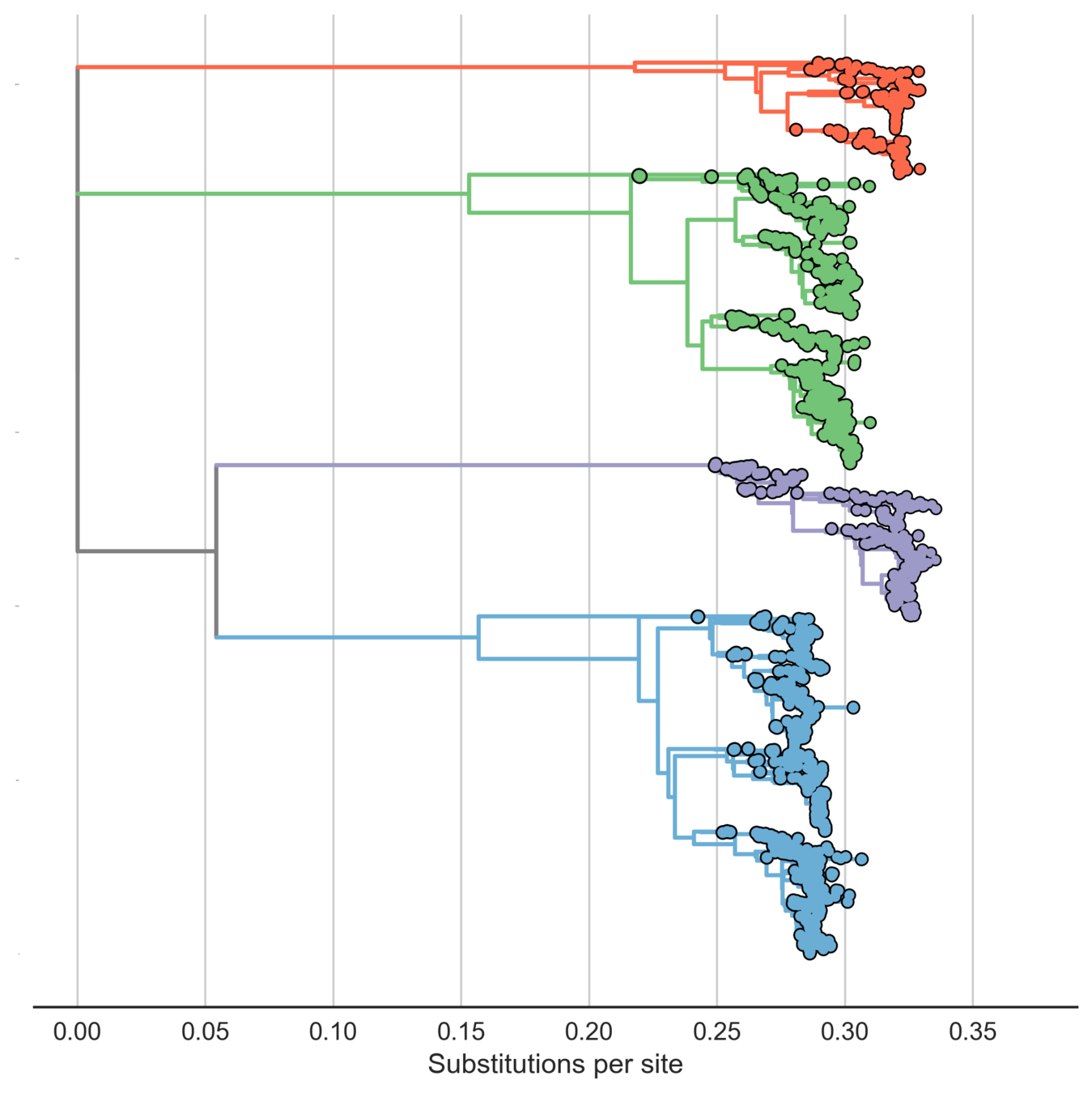
Each serotype is genetically diverse

DENV1
DENV3
DENV4
DENV2
ACTG
ACTT
AGTT



Antigenic variation
Genetic
variation
Brief detour: genetic variation can
cause "antigenic variation"

Antigenic variation changes what the virus looks like to your immune system





Measles
Antigenically uniform
Flu
Antigenically
diverse


Lifelong protection
Temporary cross-protection



Antigenic variation is
why you need a new flu shot every year
Measles is
antigenically uniform
Flu is
antigenically diverse


Dengue serotypes are antigenically distinct



Measles
Antigenically uniform
Flu
Antigenically
diverse


Within
a dengue
serotype
Antigenically uniform (?)




Between dengue serotypes
Antigenically diverse













Lifelong protection
Temporary
cross-protection
Initial
response
For dengue, antigenic differences
can cause severe disease
After
1-3 yrs



Increased risk
Dengue vaccine efficacy varies by genotype

DENV1
DENV3
DENV4
DENV2
Juraska et al., PNAS 2018
DENV antigenic relationships
are poorly understood
Serotypes are genetically distinct
Clades are genetically distinct
Serotypes are antigenically distinct
Are clades antigenically distinct?
≠
?




≠


≠


How does
dengue evolve
antigenically?
Measuring antigenic relationships


Neutralization

+






+

Replication
Neutralizing antibodies block virus replication
Titers measure antigenic distance

Antigenic similarity = how much sera is required to neutralize viral replication?
Serum



Test viruses






1x
2x
4x
(relative to autologous)
X
X
X
X
X
X



--
Log2
0
1
2
Titer
Titers measure antigenic distance



log2(titer)
antigenic
distance
cross
immunity
| 0 | 1 | 2 |
| 1 | 0 | 2 |
| 2 | 2 | 0 |
Serum
Test viruses



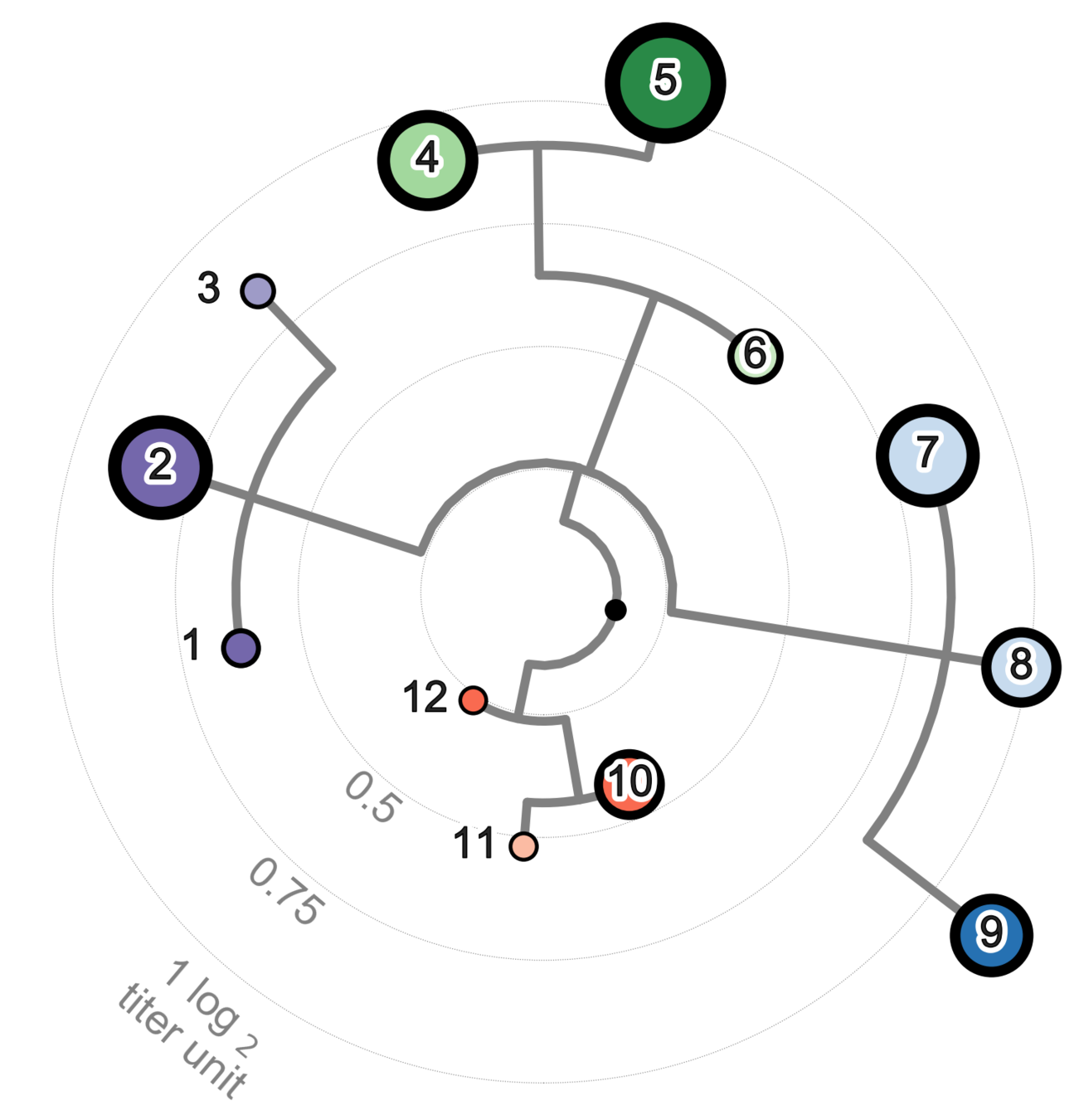
Dengue titers are confusing
Non-human primate sera; 3 months post primary infection
Katzelnick et al, Science 2015

Antigenic distance
(less cross immunity)
Cartography "maps" antigenic relationships






| 0 | 1 | 2 |
| 1 | 0 | 2 |
| 2 | 2 | 0 |



1
2
2
Antigenic Distance
log2(Titer)
Distance
Dengue serotypes look (maybe?) antigenically diverse

= 2-fold change in titer
Katzelnick et al, Science 2015
Where did this variation come from? Does it matter?
Genetic vs antigenic distance
Homotypic
Heterotypic


Phylogenies can describe both genetic and antigenic relationships

Quantify antigenic change along each branch,
Neher et al, PNAS 2016






| 0 | 1 | 2 |
| 1 | 0 | 2 |
| 2 | 2 | 0 |


Titer
between
&





+ avidity + potency

1.0
0.5
0.5
0.5
Learn these
Minimize
this
Predict titers
dTiter on each branch
avidity
potency
Predict titers


Squared Error
Minimize
this
L1 norm on branch effects
L2 norm on virus avidity & serum potency
dTiter on each branch
avidity
potency
Predict titers


Interpolate across the tree
to estimate antigenic relationships


Data
Model
Does within-serotype genetic diversity change antigenic phenotypes?
Interserotype hypothesis
Full tree hypothesis


| Pearson R | 0.79 |
| Abs. Error | 1.02 |
| 0.86 |
| 0.86 |
Between-serotype variation explains most of
dengue antigenic phenotypes
Within-serotype variation substantially contributes to dengue antigenic phenotypes
Test Error
Interserotype model
Full tree model

Bell et al., bioRxiv 2018
Each serotype contains antigenic diversity
Bell et al., bioRxiv 2018


Vaccine efficacy varies within serotypes
Juraska et al., PNAS 2018



How does
antigenic diversity
impact dengue
population dynamics?
Serotypes cycle through populations
Genotypes



Bell et al., bioRxiv 2018
Does antigenic novelty contribute to dengue clade turnover?

Population susceptibility:


Previously circulating:





Population
immunity:













Clade growth:


Does antigenic fitness
predict clade growth & decline?
Lukzsa and Lassig, Nature 2014
Growth rate @
next 5 years
How antigenically distant
is from what has recently circulated?


Does antigenic novelty contribute
to dengue fitness?
Relative frequency of j
Waning immunity
Antigenic distance between i and j
Lukzsa and Lassig, Nature, 2014
Does antigenic novelty contribute
to dengue fitness?
Relative frequency of j
Waning immunity
Probability of protection from i
given exposure to j
Lukzsa and Lassig, Nature, 2014

Serotype frequency
Predict clade growth from antigenic fitness
Antigenic novelty drives
viral fitness and serotype turnover

Pearson's r = 0.79
Bell et al., bioRxiv 2018
Measuring genotype antigenic novelty
Serotype resolution
Clade resolution


Serotype resolution
Clade resolution
Antigenic fitness contributes
to genotype turnover

Pearson's r = 0.56
Pearson's r = 0.53
Bell et al., bioRxiv 2018
Summary
2: Serotype-level antigenic relationships drive dengue
population dynamics


1: Dengue serotypes are moderately antigenic diverse
An evolutionary perspective
helps us understand how viruses...
....Enter populations
....Adapt to their hosts
....Spread through populations





+
Acknowledgements
Bedford Lab
Trevor Bedford, Alli Black, Gytis Dudas, James Hadfield, John Huddleston, Katie Kistler, Maya Lewinsohn, Colin Megill, Louise Moncla, Barney Potter, Tom Sibley
Collaborators & Colleagues
Sarah Cobey, Michael Emerman, Jeremy Freeman, Ted Gobillot, Leah Katzelnick, Richard Neher, Molly O'hainle, Duncan Ralph, David Shaw, Frank Wen, Janet Young
Committee
Jesse Bloom, Julie Overbaugh, Adam Geballe, Erick Matsen, & Daniela Witten
Computational Biology
Melissa Alvendia, Sam Distel, Lindsey Mwoga, An Tyrrell, Scientific Computing
Graduate Program
Rich Gardner, Katie Peichel & Nina Salama
Andrea Brocato, Denise Barnes, Tierra Johnson, Maia Lowe, Laura Masserman, Teresa Swanson
Outreach
Lori Blake, Jeanne Chowning, Sarah Hilton, Emma Hoppe, Shannan Moran, Liza Ray, Beverly Torok-Storb, Kim Wells
Funding
NSF Graduate Research Fellowship, NIH Interdisciplinary Training Grant, ARCS & Vassar Scholar Awards
Slides: RevealJS & The Noun Project
Questions?
@sidneymbell


slides.com/sidneymbell/dissertation