Disseny i aplicació de bases de dades biològiques per a la classificació i anotació de proteïnes
Directors:
Enric Querol
Francesc Xavier Avilés
Objectius
-
Posar al dia diferents aplicacions bioinformàtiques desenvolupades anys abans a l'IBB-UAB.
- Per utilitzar-les en llocs i serveis web accessibles per a tota la comunitat científica.
- Per acoblar-les a diferents processos d'anàlisi massius (resultat dels projectes de seqüenciació de genomes).
- Dissenyar repositoris de coneixement (bases de dades) que permetin accedir als anàlisis resultants de les anteriors aplicacions i altres.
Objectius
- Implementar un entorn d'anàlisis de seqüències per tal que eines populars de tercers, com ara el BLAST, com les anteriorment esmentades desenvolupades a l'IBB, puguin utilitzar-se sense necessitat de grans coneixements tècnics.
- Recerca i determinació funcional, estructural i filogenètica de nous membres de la família de les proteïnes metal·locarboxipeptidades.
Dades biològiques
Modelització biològica


A
Formats bioinformàtics
FASTA
>sp|Q9BTV4|TMM43_HUMAN Transmembrane protein
MAANYSSTSTRREHVKVKTSSQPGFLERLSETSGGMFVGLMAFLLSFYLIFTNEGRALKT
ATSLAEGLSLVVSPDSIHSVAPENEGRLVHIIGALRTSKLLSDPNYGVHLPAVKLRRHVE
MYQWVETEESREYTEDGQVKKETRYSYNTEWRSEIINSKNFDREIGHKNPSAMAVESFMA
TAPFVQIGRFFLSSGLIDKVDNFKSLSLSKLEDPHVDIIRRGDFFYHSENPKYPEVGDLR
VSFSYAGLSGDDPDLGPAHVVTVIARQRGDQLVPFSTKSGDTLLLLHHGDFSAEEVFHRE
LRSNSMKTWGLRAAGWMAMFMGLNLMTRILYTLVDWFPVFRDLVNIGLKAFAFCVATSLT
LLTVAAGWLFYRPLWALLIAGLALVPILVARTRVPAKKLEFormats bioinformàtics
PDB
ATOM 1 N HIS A 1A 16.838 -16.537 -15.237 1.00 58.22 N
ATOM 2 CA HIS A 1A 15.999 -17.073 -14.177 1.00 57.61 C
ATOM 3 C HIS A 1A 15.313 -15.951 -13.398 1.00 56.85 C
ATOM 4 O HIS A 1A 15.739 -14.799 -13.451 1.00 54.65 O
ATOM 5 CB HIS A 1A 16.818 -17.936 -13.211 1.00 59.70 C
ATOM 6 CG HIS A 1A 18.008 -17.208 -12.662 1.00 61.33 C
ATOM 7 ND1 HIS A 1A 19.282 -17.394 -13.150 1.00 62.30 N
ATOM 8 CD2 HIS A 1A 18.126 -16.283 -11.684 1.00 61.06 C
ATOM 9 CE1 HIS A 1A 20.127 -16.621 -12.488 1.00 62.16 C
ATOM 10 NE2 HIS A 1A 19.450 -15.935 -11.584 1.00 61.36 N
ATOM 11 N HIS A 2A 14.260 -16.335 -12.694 1.00 55.96 N
ATOM 12 CA HIS A 2A 13.485 -15.490 -11.802 1.00 55.45 C
ATOM 13 C HIS A 2A 12.710 -14.403 -12.538 1.00 49.55 C
ATOM 14 O HIS A 2A 12.408 -13.349 -11.974 1.00 47.18 O
ATOM 15 CB HIS A 2A 14.410 -14.891 -10.737 1.00 61.53 C
ATOM 16 CG HIS A 2A 14.735 -15.881 -9.655 1.00 66.23 C
ATOM 17 ND1 HIS A 2A 13.775 -16.384 -8.802 1.00 67.88 N
ATOM 18 CD2 HIS A 2A 15.895 -16.458 -9.272 1.00 68.10 C
ATOM 19 CE1 HIS A 2A 14.331 -17.228 -7.947 1.00 69.31 C
ATOM 20 NE2 HIS A 2A 15.625 -17.291 -8.214 1.00 69.65 N
ATOM 21 N GLY A 3A 12.370 -14.682 -13.795 1.00 44.60 N
ATOM 22 CA GLY A 3A 11.616 -13.760 -14.621 1.00 41.29 C
ATOM 23 C GLY A 3A 12.481 -12.660 -15.198 1.00 37.90 C
ATOM 24 O GLY A 3A 12.002 -11.758 -15.886 1.00 35.74 O
ATOM 25 N GLY A 4A 13.782 -12.724 -14.927 1.00 35.35 N
ATOM 26 CA GLY A 4A 14.672 -11.669 -15.390 1.00 33.87 C
ATOM 27 C GLY A 4A 14.604 -11.547 -16.899 1.00 33.53 C
ATOM 28 O GLY A 4A 14.757 -10.459 -17.447 1.00 35.80 O
ATOM 29 N GLU A 5A 14.379 -12.668 -17.579 1.00 35.10 N
ATOM 30 CA GLU A 5A 14.368 -12.630 -19.040 1.00 37.50 C
ATOM 31 C GLU A 5A 13.352 -11.623 -19.568 1.00 37.65 C
ATOM 32 O GLU A 5A 13.587 -10.974 -20.594 1.00 37.89 O 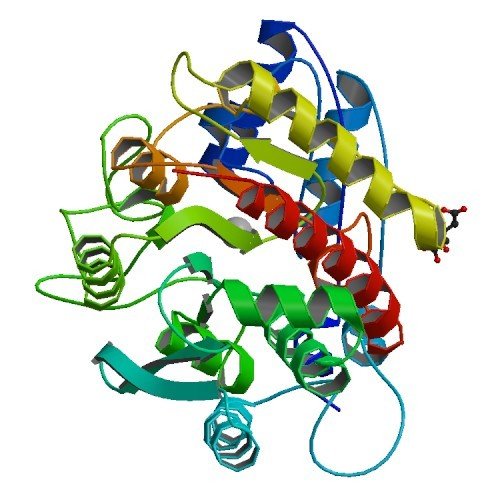
Formats bioinformàtics
GFF

Dogma central
Font: Commons
##gff-version 3
#!genome-build ASM2734v1
#!genome-version GCA_000027345.1
#!genome-date 2014-01
#!genome-build-accession GCA_000027345.1
#!genebuild-last-updated 2014-01
Chromosome ena gene 380541 381527 . + . ID=MPN_320;Name=thyA
Chromosome ena mRNA 380541 381527 . + . ID=AAB96164;Name=thyA-1;Parent=MPN_320
Chromosome ena start_codon 380541 380543 . + 0 ID=start_codon:AAB96164:1;Parent=AAB96164
Chromosome ena CDS 380541 381527 . + 0 ID=CDS:AAB96164:1;Parent=AAB96164
Chromosome ena transcript 380541 381527 . + . ID=transcript:AAB96164:1;Parent=AAB96164
Chromosome ena exon 380541 381527 . + . ID=exon:AAB96164:1;Parent=AAB96164
Chromosome ena stop_codon 381525 381527 . + 0 ID=stop_codon:AAB96164:1;Parent=AAB96164
Chromosome ena gene 293265 294107 . + . ID=MPN_241;Name=MPN_241
Chromosome ena mRNA 293265 294107 . + . ID=AAB96238;Parent=MPN_241
Chromosome ena start_codon 293265 293267 . + 0 ID=start_codon:AAB96238:1;Parent=AAB96238
Chromosome ena CDS 293265 294107 . + 0 ID=CDS:AAB96238:1;Parent=AAB96238
Chromosome ena exon 293265 294107 . + . ID=exon:AAB96238:1;Parent=AAB96238
Chromosome ena transcript 293265 294107 . + . ID=transcript:AAB96238:1;Parent=AAB96238
Chromosome ena stop_codon 294105 294107 . + 0 ID=stop_codon:AAB96238:1;Parent=AAB96238
Chromosome ena gene 760403 760948 . + . ID=MPN_634;Name=MPN_634
Chromosome ena mRNA 760403 760948 . + . ID=AAB95856;Parent=MPN_634
Chromosome ena start_codon 760403 760405 . + 0 ID=start_codon:AAB95856:1;Parent=AAB95856
Chromosome ena CDS 760403 760948 . + 0 ID=CDS:AAB95856:1;Parent=AAB95856
Chromosome ena transcript 760403 760948 . + . ID=transcript:AAB95856:1;Parent=AAB95856
Chromosome ena exon 760403 760948 . + . ID=exon:AAB95856:1;Parent=AAB95856
Chromosome ena stop_codon 760946 760948 . + 0 ID=stop_codon:AAB95856:1;Parent=AAB95856
Chromosome ena gene 773342 773752 . - . ID=MPN_648;Name=MPN_648
Chromosome ena mRNA 773342 773752 . - . ID=AAB95842;Parent=MPN_648
Chromosome ena start_codon 773750 773752 . - 0 ID=start_codon:AAB95842:1;Parent=AAB95842
Chromosome ena CDS 773342 773752 . - 0 ID=CDS:AAB95842:1;Parent=AAB95842
Chromosome ena transcript 773342 773752 . - . ID=transcript:AAB95842:1;Parent=AAB95842
Chromosome ena exon 773342 773752 . - . ID=exon:AAB95842:1;Parent=AAB95842
Chromosome ena stop_codon 773342 773344 . - 0 ID=stop_codon:AAB95842:1;Parent=AAB95842
Chromosome ena gene 728749 729738 . - . ID=MPN_609;Name=pstBFormats NO bioinformàtics
P. ex. full de càlcul amb dades diverses

Bases de dades relacionals
Esquema d'una taula

- Atribut, Camp, Columna
- Tupla, fila
- Taula, relació
Relacions entre taules

Esquema d'una BD
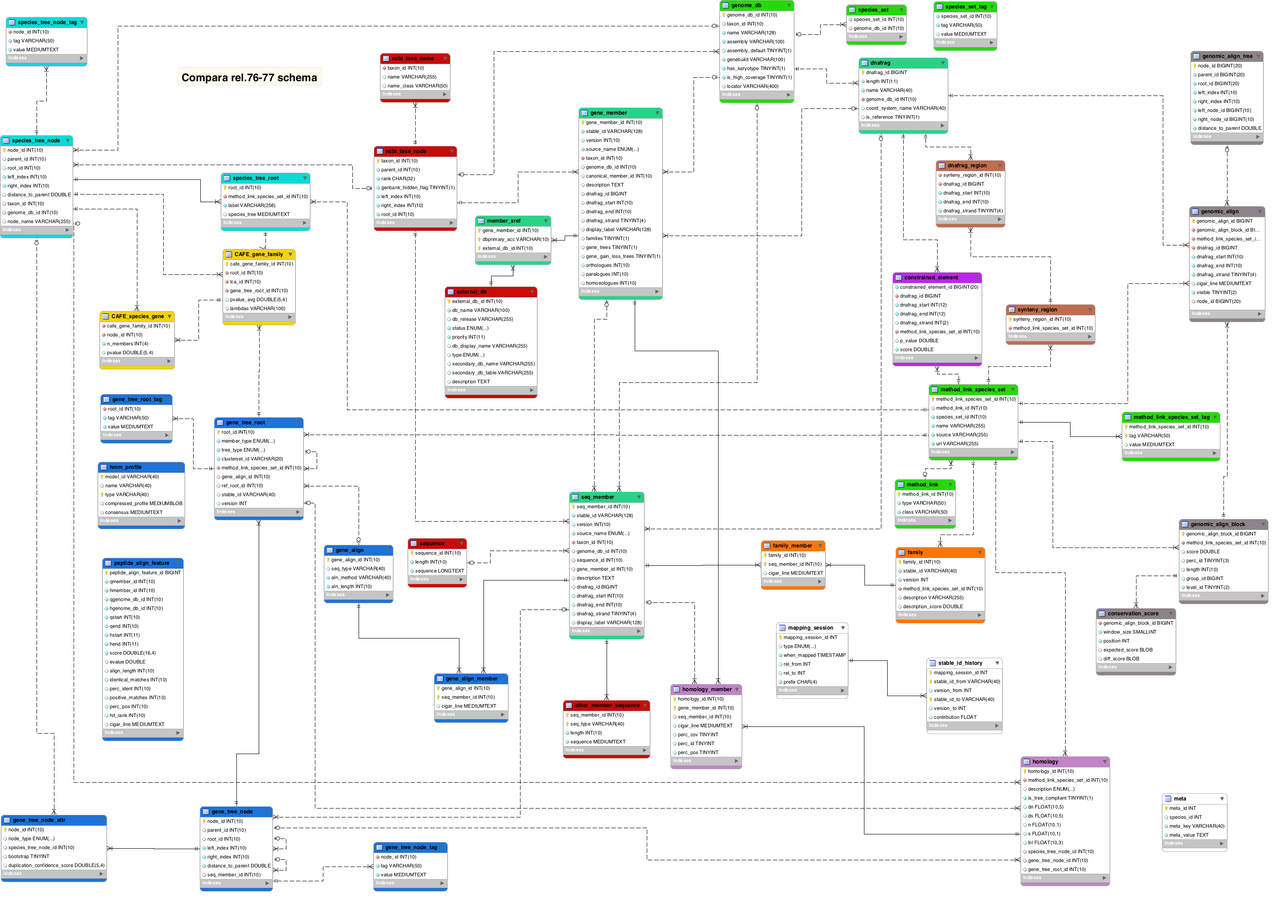
Esquema d'ENSEMBL Compara
SQL
Llenguatge de consulta estructurat
SQL - Definició de taula
CREATE TABLE `translation` (
`translation_id` int(10) unsigned NOT NULL AUTO_INCREMENT,
`transcript_id` int(10) unsigned NOT NULL,
`seq_start` int(10) NOT NULL,
`start_exon_id` int(10) unsigned NOT NULL,
`seq_end` int(10) NOT NULL,
`end_exon_id` int(10) unsigned NOT NULL,
`stable_id` varchar(128) DEFAULT NULL,
`version` smallint(5) unsigned NOT NULL DEFAULT '1',
`created_date` datetime NOT NULL DEFAULT '0000-00-00 00:00:00',
`modified_date` datetime NOT NULL DEFAULT '0000-00-00 00:00:00',
PRIMARY KEY (`translation_id`),
KEY `transcript_idx` (`transcript_id`),
KEY `stable_id_idx` (`stable_id`,`version`)
);SQL - SELECT
SELECT translation_id, seq_start, seq_end
from translation
where transcript_id='ENST00000296026';Aplicacions bioinformàtiques
ProtLoc
(Cedano et al., 1997)
CODE
ProtLoc

Localitzacions cel·lulars
Source - NSF - Commons: How proteins are made?
ProtLoc - Premisses
- Assignar una ubicació cel·lular només amb la seqüència primària d'una proteïna
- Localitzacions assignables:
- INTRACEL·LULAR
- EXTRACEL·LULAR
- MEMBRANA
- ANCORADA
- NUCLEAR
ProtLoc
Distància de Mahalanobis



ProtLoc
Resultats
Membrane => 8.04007866311015 Anchored => 8.70351093081404 Nuclear => 8.92771314050655 Intracellular => 9.33241032405688 Extracellular => 11.0166179413074
TMM43_HUMAN (Transmembrane protein 43)
Aplicacions bioinformàtiques
TransMem
CODE
(Aloy et al., 1997)
TransMem
Dominis transmembrana

TransMem - Premisses
- Assignar dominis transmembrana només amb la seqüència primària d'una proteïna
- Ajustament de la finestra de detecció - ref.: 20 aa per hèlix transmembrana
TransMem

SNNS (Stuttgart Neural Network Simulator)
Xarxa Neuronal Artificial
TransMem
Resultats
$ ./transmem -w 10 Q9BTV4.fasta
sp|Q9BTV4|TMM43_HUMAN GMFVGLMAFLLSFYL 34 49
sp|Q9BTV4|TMM43_HUMAN VESFMATAPF 174 184
sp|Q9BTV4|TMM43_HUMAN IGRFFLSSGL 186 196
sp|Q9BTV4|TMM43_HUMAN AAGWMAMFMGLNLM 312 326
sp|Q9BTV4|TMM43_HUMAN LVNIGLKAFAFCVATSLTLLTVAAGWLF 342 370
sp|Q9BTV4|TMM43_HUMAN LWALLIAGLALVPIL 373 388
Transmembrane protein 43
Aplicacions bioinformàtiques
TranScout
(Aguilar et al., 2002)
TranScout
Factors de transcripció

Source: Commons - Leucine Zipper
TranScout - Premisses
- Detecció de diferents tipus de factors de transcripció a partir de diferents perfils de seqüència
-
Exemple de tipus:
- Bzip-domains
- Homeodomains
- Cys4 Zn fingers (metall)
- etc.
- Detall: http://bioinf.uab.es/transcout/class.html
TranScout
Perfils de seqüència

TranScout
Matriu de puntuació específica de posició (PSSM)

Exemple de PSSM de tipus zinc finger
TranScout
Resultat
QUERY KRPRTAFTAEQ-LQRLKNEFQNNRYLTEQRR-QALAQELGLNESQIKIWFQNKRAKIK
MOTIF pphpphhpppphhp-hppphphNpYLpRpRRhE-hAphLpLpERQhKIhhpppphphp
INICIO 173
FINAL 228
SIGN Antennapedia type of homeotic transcription factors
CLASS Family
FUNC DNA-binding
SCORE 22.93
SCRNRM 0.40
SCRMAX 33.91
SCRMIN 16.64
RANK PROBABLE
Aplicacions de bases de dades en biologia
Referència del sector - NAR DB issue: http://www.oxfordjournals.org/nar/database/c/
Basades en els programes anteriors
TrSDB
Base de dades de factors de transcripció en eucariotes
(Hermoso et al., 2004)
Fonts i programes que s'han utilitzat
- IPI i SwissProt-TrEMBL --> Avui UniProt
- InterPro (aglutina diferents BDs)
- InterPro2GO - GOA -> Anotació funcional
- ProtLoc
- TransMem
- TranScout
Perspectiva

Perspectiva

Resultats

Resultats

ArchDB
Base de dades de llaços de proteïnes
(Espadaler et al., 2004)
(Hermoso et al., 2007)
Paràmetres per a la classificació de llaços
- Assignació estructura secundària
- Conformació phi-psi (Ramachandran)
- Geometria (angles i distància) dels llaços
Mapa de Ramachandran
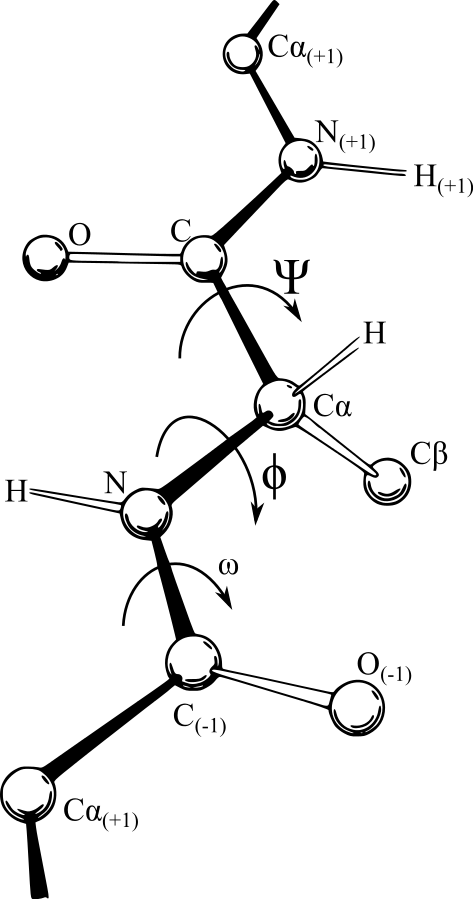

Source: Commons
Source: Commons
Geometria dels llaços de proteïnes

- rho
- delta
- theta
- D
Flux de l'ArchDB

Classificació
Perspectiva

Perspectiva

Plataforma d'anotació de proteïnes
Objectius
- Enviar dades biològiques (seqüències, pero potencialment també altres)
- Gestionar-les (tenir-les) en un mateix lloc
- Aplicar-les-hi anàlisis i relacionar-ne els resultats
NoSQL
NoSQL
Not Only SQL
- No només SQL
- Sistemes de bases de dades no relacionals
- Molts tipus:
- Clau-valor
- Documents
- Grafs
- Mixtes
- etc.

Bases de dades de documents
CouchDB
Què entenem a CouchDB per document?
- Unitats diferenciades de dades (p. ex., 1 anàlisi de BLAST)
-
Subjectes a operacions CRUD
- Create (Crear)
- Read (Llegir)
- Update (Actualitzar)
- Delete (Suprimir)
- Amb una clau unívoca
- Revisions
- Diferents grups (i jerarquies) de dades
Document

Resultat XML de BLAST
Document JSON

Un mateix resultat de BLAST
Document en CouchDB

Resultat de Bypass
Característiques respecte al model de BD relacional
- Sense un esquema predefinit
- El document determina l'esquema i cada document en pot tenir un diferent
- El model passa a nivell de programari
Bases de dades
de grafs
Neo4j
Grafs

- Vèrtexs o nodes
- Arestes o relacions
- Propietats o atributs
- Etiquetes
Els grafs explicats
amb un graf
Tipus de grafs

Jerarquia simple. Regla directa.
1 pare.
Taxonomia NCBI.
Graf acíclic directe. Regles directes, però >= 1 pare.
Gene Ontology (3).
Graf no directe. Els nodes poden ser tan pares com fills.
Xarxes metabòliques.
Graf taxonomia NCBI

Visualització Neo4j
Ontologia component cel·lular del GO

Mapa d'interacció de proteïnes de llevat

Ancestre comú més baix

Exemple: LCA GO de proteïnes

Bypass
Reanotació funcional basada
en lògica difusa
(Gómez et al., 2008)
BLAST

BLAST i PSI-BLAST

Moonlighting
- Moonlighting: de l'anglès, treballar en una altra feina apart de la principal (normalment durant la nit) - Escena Taxi Driver
- Biomolècules amb una funcionalitat addicional a una canònica
- Relacionat: promiscuïtat enzimàtica
Lògica difusa

Font: Commons
Paràmetres del Bypass
-
Composició d'aminoàcids
- Distància de dissimilaritat euclidiana del conjunt
- Longitud de la seqüència
-
Perfil hidropàtic
- Kyte-Doolittle
-
Perfil de flexibilitat
- Karplus & Schulz
Flux del Bypass

Reordenació de resultats de PSI-BLAST

Exemples de moonlighting coincidents amb resultats de Bypass i interactòmica
Més exemples a MultitaskProtDB

(Hernández et al., 2014)
Integració en plataforma
Entrada de dades i anàlisi

Reordenació amb Bypass

Integració amb API GO

Futures implementacions
- Importació i exportació més fàcil (ara mitjançant scripts)
- Integrar més microserveis WWW ( TransMem, ProtLoc, altres externs)
- Major control de la creació del perfil de PSSM del PSI-BLAST
- Parametritzar millor el Bypass
M14D
Una 'jove' subfamília de metal·locarboxipeptidases
Metal·locarboxipeptidases
-
Exopeptidases. Tallen extrem C-terminal pèptids
- En contraposició a aminopeptidases i endopeptidases
-
El centre actiu té un metall (majoritàriament zinc).
- Altres carboxipeptidases ho fan mitjançant serina o cisteïna.
Metal·locarboxipeptidases
- Alguns membres (especialment si s'excreten fora de la cel·lula) tenen una forma inactiva (zimògen/proenzim) amb un domini protector.
- Família M14 al Merops - la base de dades de referència de peptidases
Metal·locarboxipeptidases


M14A - Procarboxipeptidasa B humana
M14B - Carboxipeptidasa N humana
Metal·locarboxipeptidases
Residus rellevants

Variacions en els residus d'unió a zinc en les diferents famílies de metal·locarboxipetidases
Recerca de nous membres de la família M14
(Rodriguez de la Vega et al., 2007)
(Kalinina et al., 2007)
TubCP =? Nna1 =? M14D (CCP)
- La literatura es referia fa temps a una activitat tubulin-carboxipeptidasa al citosol, però d'una proteïna indeterminada
- Nna1 - associat a desordre neuronal (pcd en ratolí - degeneració de cel·lules de Purkinje)
- Suposadament també alguns membres amb possibilitat d'unir ATP/GTP (AGTPBP1 en humans )
Generació d'alineaments

Origen: Merops M14D
Perfil amb un model ocult de Markov

Diversitat de la subfamília

Les 6 carboxipeptidases citosòliques (CCP - M14D) de ratolí.
Font: (Kalinina et al., 2007)
Diversitat de la subfamília

Font: (Rodriguez de la Vega et al., 2007)
Arbre filogenètic

Font: (Rodriguez de la Vega et al., 2007)
Modelització estructural
Modelització per homologia
- Utilitzat MODELLER (Sali Lab)
-
Procediment:
- Es parteix d'unes estructures 3D ja existents (PDB)
- Es creen alineaments entre les seqüències de les estructures i la nostra seqüència problema
- ->
Modelització per homologia
Preparació d'alienaments per a MODELLER
>P1;1aye
structure:1aye:34C : :11 : :33-401::-1.00:-1.00
QLDFWKSPTT--------PGETAHVRVPFVNVQAVKVFLESQGIAYSIMIEDVQVLLDKENEEMLFNRRRERSG
NFNFGA
---------------------------------------------------------------------
--------------------------------------------------------------------------
--------------------------------------------------------------------------
--------------------------------------------------------------------------
-------*
>P1;1h8lA
structure:1h8l:7 :A:383 :A:4-380::-1.00:-1.00
--------------------------------------------------------------------------
QPVDFRHHHFSDMEIFLRRYANEYPSITRLYSVGKSVELRELYVMEISDNPGIHEAGEPEFKYIGNMHGNEVVGR
ELLLNL
IEYLCKNFGTDPEVTDLVQSTRIHIMPSMNPDGYEKSQEGDRGGTVGRNNSNNYDLNRNFPDQFFQVTD
PPQPETLAVMSWLKTYPFVLSANLHGGSLVVNYPFDDDEQGI-----AIYSKSPDDAVFQQLALSYSKENKKMYQ
GSPCKDLYPTEYFPHGITNGAQWYNVPGGMQDWNYLNTNCFEVTIELGCVKYPKAEELPKYWEQNRRSLLQFIKQ
VHRGIWGFVLDATDGRGILNATISVADINHPVTTYKDGDYWRLLVQGTYKVTASARGYDPVTKTVEVDSKGGVQV
NFTLSRT*
>P1;CPM
sequence:model: : : : :CPM 10-462::-1.00:-1.00
HSQTHRRPTMILRTPLVAGHQLQLLLLPSVLLLMLHLLLCDAKTVNPGDSMQMQHHQMTAEPGLPEPRAYMPDA
QHLDFV
YHDHEELTRFLRATSARYPNLTALYSIGKSIQGRDLWVMVVSSSPYEHMVGKPDVKYVGNIHGNEPVGR
EMLLHLIQYFVTSYNTDQYVKWLLDNTRIHILPTMNPDGYAVSKEGTCDGGQGRYNARGFDLNRNFPDYFKQNNK
RGQPETDSVKDWISKIQFVLSGSLHGGALVASYPYDNTPNSMFQTYSAAPSLTPDDDVFKHLSLVYARNHAKMSR
GVACKSA--TPAFENGITNGAAWYPLTGGMQDYNYVWYGCMEITLEISCCKFPPAYELKKYWEDNQLSLIKFLAE
AHRGVQGFVFDPA-GMPIERASIKIKGRDVGFQTTKYGEFWRILLPGYYKVEVFAEGFAPREVEFVIVEQHPTLL
NVTLQPS*Exemple d'aplicació

Model de M14D - Homòleg d'AGBL4 a C. elegans (ceAGBL4). Plantilles: CPA pancreàtica de Bos taurus (2ctc) i Sus scrofa (1pca), carboxipeptidasa pancreàtica B (CPB) de Sus scrofa (1nsa) i CPT de Thermoactinomyces vulgaris (1obr)
Font: (Rodriguez de la Vega et al., 2007)
Estat actual CCP - M14D
-
CCP1 en ratolí (cpd) demostrat que intervé en el processament Glu de la α- i β-tubulina (Berezniuk et al., 2012)
- Membres clau en el processament de microtúbuls (regeneració axons, etc.)
-
Estructura de CCP de bacteri Pseudomonas aeruginosa - PaCCP (Otero et al., 2012)
- Nou domini pro
- Possibilitats per a millors modelitzacions
Conclusions finals
-
Es van reescriure, adaptar o encapsular diferents aplicacions (ProtLoc, TransMem, TranScout i Bypass).
- Van poder utilitzar-se en processament massiu de dades dels projectes genoma.
- Van poder exposar-se al públic com a llocs web.
- Esdevé possible integrar-les com a serveis web per a ser utilitzades en entorns WWW.
Conclusions finals
-
Es van dissenyar bases de dades de coneixement d'accés públic d'entitats d'interès biològic.
- TrSDB. Repositori de factors de transcripció coneguts i putatius a partir dels anàlisis amb dades públiques i les aplicacions abans esmentades.
- ArchDB. Repositori de llaços de proteïnes categoritzats segons criteris estructurals, geomètrics i funcionals.
Conclusions finals
-
Es va implementar un entorn d'anàlisi funcional de seqüències de proteïnes.
- Aglutinant anàlisis de (PSI-)BLAST, Bypass i altres aplicacions.
- Experimentant amb tecnologies NoSQL per a l'emmagatzemament de les dades.
- Permetent a l'usuari valorar una anotació mitjançant la integració amb Gene Ontology i taxonomia via microserveis web.
Conclusions finals
-
Es va determinar una nova subfamília de metal·locarboxipeptidades (M14D)
- Mitjançant cerca en bases de dades públiques van trobar-se representants en gran nombre d'organismes i es va proposar una història filogenètica de tot el grup.
- Es van generar models estructurals per donar suport a una possible funció catalítica.
- En definitiva, es va obrir un camp de recerca; confirmant-se després la importància en processos cabdals com el processament de microtúbuls.
Publicacions
Hermoso, A., Aguilar, D., Aviles, F. X., & Querol, E. (2004). TrSDB: a proteome database of transcription factors. Nucleic Acids Research, 32(Database issue), D171–D173.
Espadaler, J., Fernandez-Fuentes, N., Hermoso, A., Querol, E., Aviles, F. X., Sternberg, M. J. E., & Oliva, B. (2004). ArchDB: automated protein loop classification as a tool for structural genomics. Nucleic Acids Research, 32(Database issue), D185–D188.
Hermoso, A., Espadaler, J., Enrique Querol, E., Aviles, F. X., Sternberg, M. J. E., Oliva, B., … Fernandez-Fuentes, N. (2007). Including Functional Annotations and Extending the Collection of Structural Classifications of Protein Loops (ArchDB). Bioinformatics and Biology Insights, 1, 77.
Publicacions
Gómez, A., Cedano, J., Espadaler, J., Hermoso, A., Piñol, J., & Querol, E. (2008). Prediction of protein function improving sequence remote alignment search by a fuzzy logic algorithm. Protein Journal, 27(2), 130–139.
Hernández, S., Calvo, A., Ferragut, G., Franco, L., Hermoso, A., Amela, I., … Cedano, J. (2014). Can bioinformatics help in the identification of moonlighting proteins? Biochemical Society Transactions, 42(6), 1692–7.
Hernández, S., Franco, L., Calvo, A., Ferragut, G., Hermoso, A., Amela, I., … Cedano, J. (2015). Bioinformatics and Moonlighting Proteins. Frontiers in Bioengineering and Biotechnology, 3, 90.
Publicacions
Rodriguez de la Vega, M., Sevilla, R. G., Hermoso, A., Lorenzo, J., Tanco, S., Diez, A., … Avilés, F. X. (2007). Nna1-like proteins are active metallocarboxypeptidases of a new and diverse M14 subfamily. FASEB Journal, 21(3), 851–865
Kalinina, E., Biswas, R., Berezniuk, I., Hermoso, A., Aviles, F. X., & Fricker, L. D. (2007). A novel subfamily of mouse cytosolic carboxypeptidases. FASEB Journal, 21(3), 836–50.
Gràcies a








