Reproducible BioCompute Objects using Common Workflow Language
Stian Soiland-Reyes
eScience lab, The University of Manchester
BioCompute Objects Proof of Concept Workshop
Washington DC, 2018-03-23
This work is licensed under a
Creative Commons Attribution 4.0 International License.





a metagenomics case study

Findable
Accessible
Interoperable
Reusable

To be Findable:
F1. (meta)data are assigned a globally unique and persistent identifier
F2. data are described with rich metadata (defined by R1 below)
F3. metadata clearly and explicitly include the identifier of the data it describes
F4. (meta)data are registered or indexed in a searchable resource
To be Accessible:
A1. (meta)data are retrievable by their identifier using a standardized communications protocol
A1.1 the protocol is open, free, and universally implementable
A1.2 the protocol allows for an authentication and authorization procedure, where necessary
A2. metadata are accessible, even when the data are no longer available
To be Interoperable:
I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation.
I2. (meta)data use vocabularies that follow FAIR principles
I3. (meta)data include qualified references to other (meta)data
To be Reusable:
R1. meta(data) are richly described with a plurality of accurate and relevant attributes
R1.1. (meta)data are released with a clear and accessible data usage license
R1.2. (meta)data are associated with detailed provenance
R1.3. (meta)data meet domain-relevant community standards
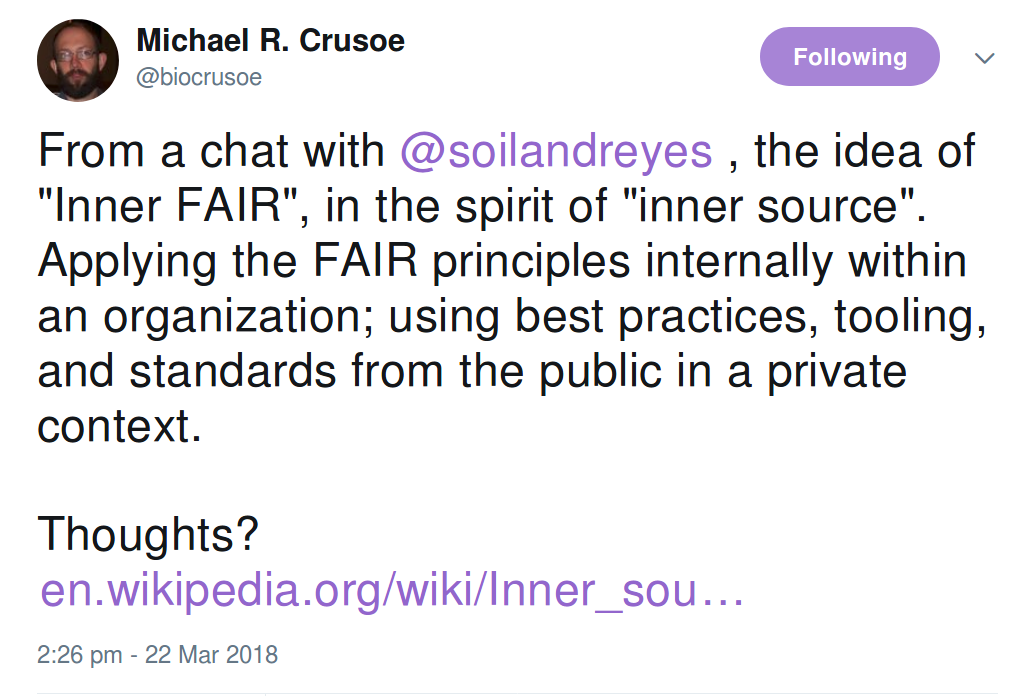
Inner source
Use of open source software development best practices and the establishment of an open source-like culture within organizations.
The organization may still develop proprietary software, but internally opens up its development.
Inner FAIR

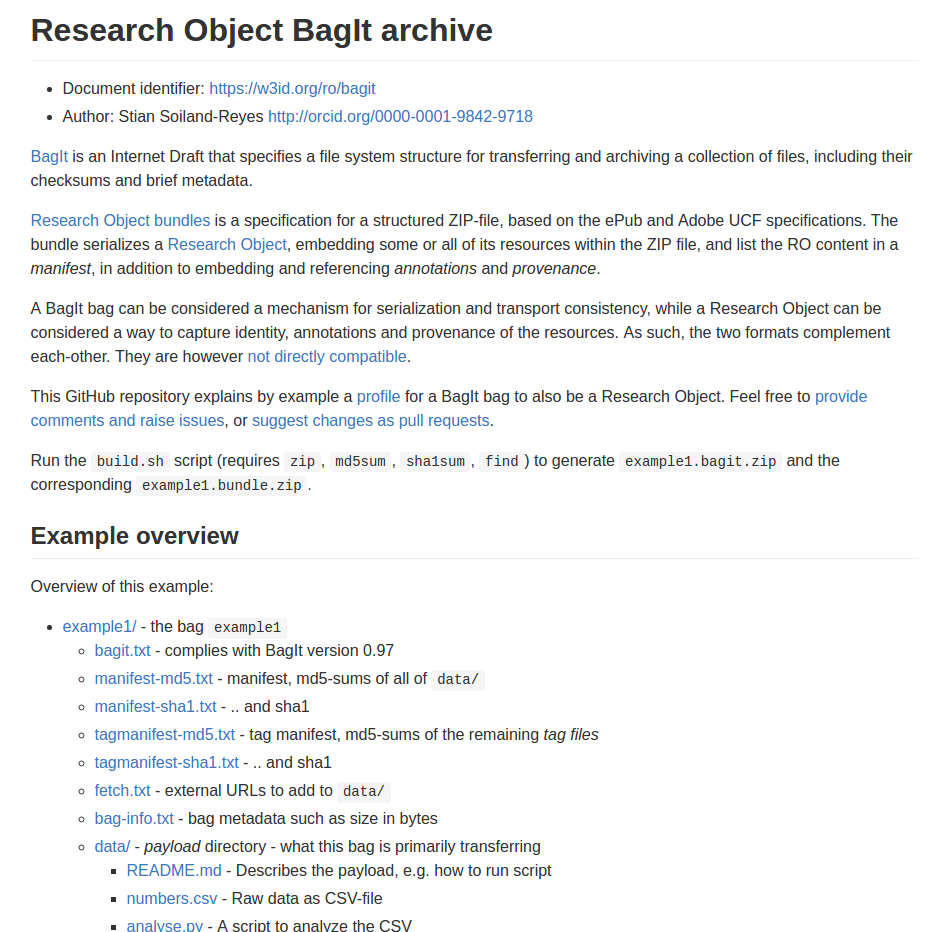
id: doi:10.15490/seek.1.investigation.56
createdOn: 2015-07-10T16:46:00Z
createdBy: http://orcid.org/0000-0001-9842-9718
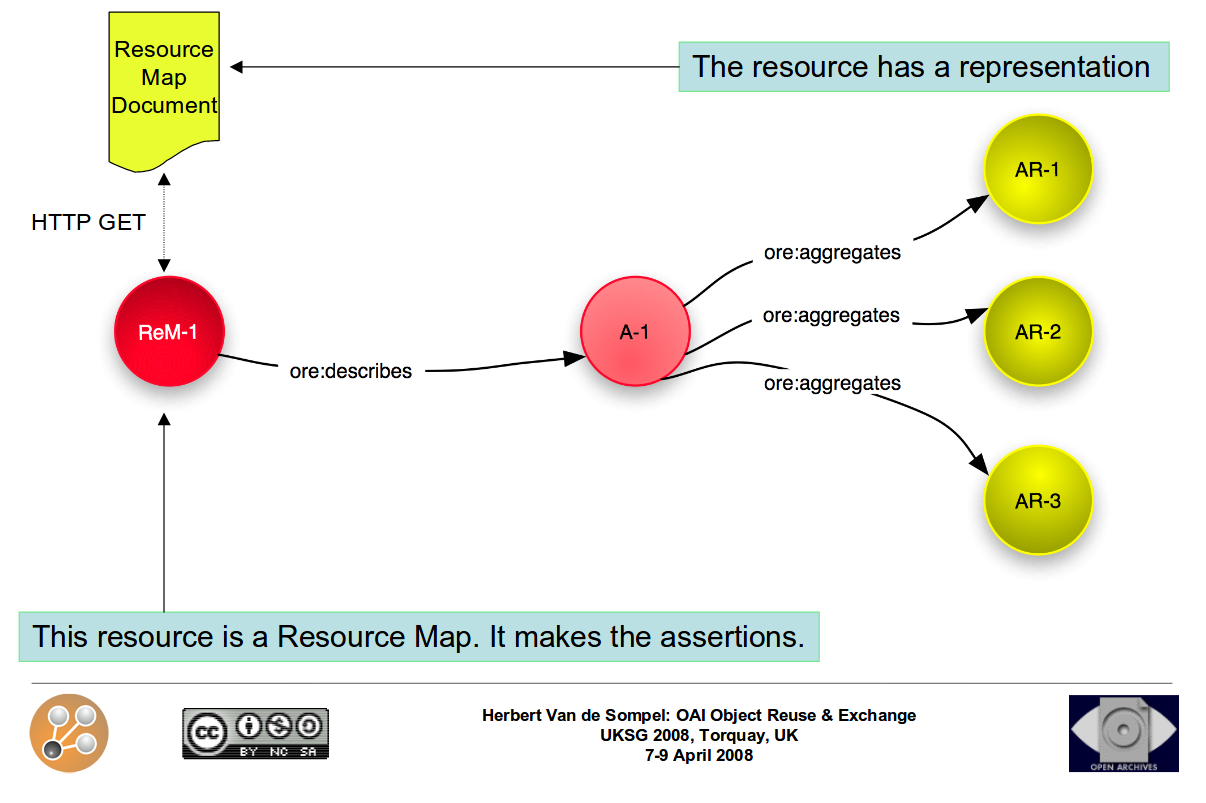
aggregates:
- id: data/sequence/specimen5.bam
conformsTo: http://gemrb.org/iesdp/file_formats/ie_formats/bam_v1.htm
- id: http://example.com/blog/about-specimen5
authoredBy: http://orcid.org/0000-0001-7066-3350
- id: http://www.myexperiment.org/workflows/3355
history: provenance/workflow-evolution.ttl
annotations:
- about: data/sequence/specimen5.bam
content: annotations/specimen5-properties.jsonld
createdBy: http://orcid.org/0000-0001-7066-3350
- about: data/sequence/specimen5.bam
content: http://example.com/blog/about-specimen5
motivatedBy: oa:questioningResearch Object manifest
(simplified)
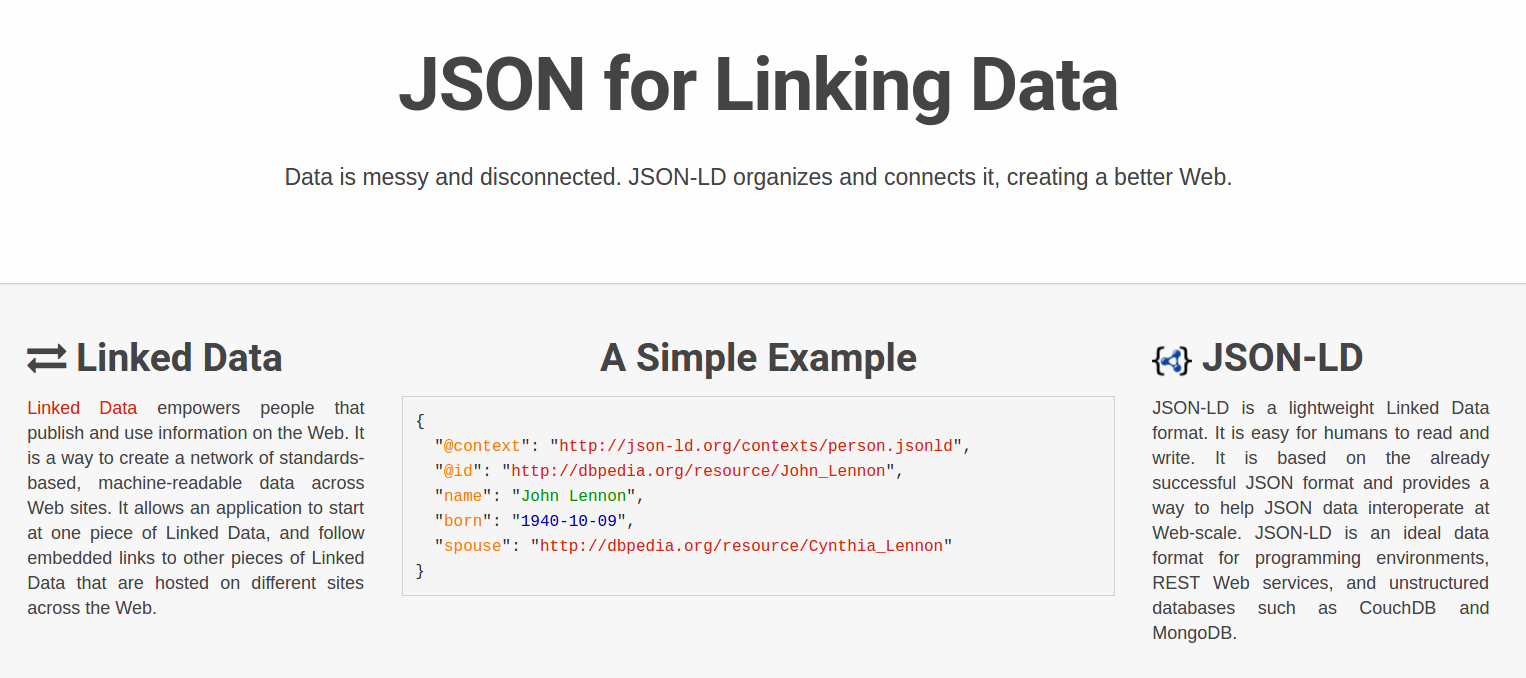
Reuse standards:
OAI-ORE, BagIt, W3C JSON-LD, PROV, Web Annotation Model

metadata/manifest.json
data/sequence/specimen5.bam
provenance/workflow-evolution.ttl
http://example.com/blog/about-specimen5
http://www.myexperiment.org/workflows/335
http://orcid.org/0000-0001-7066-3350
http://gemrb.org/iesdb/
file_formats_ie_formats_bam_v1.html









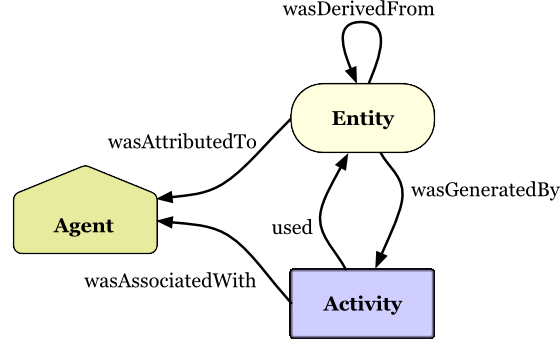
Copyright © 2013 W3C® (MIT, ERCIM, Keio, Beihang), All Rights Reserved.
W3C liability, trademark and document use rules apply.
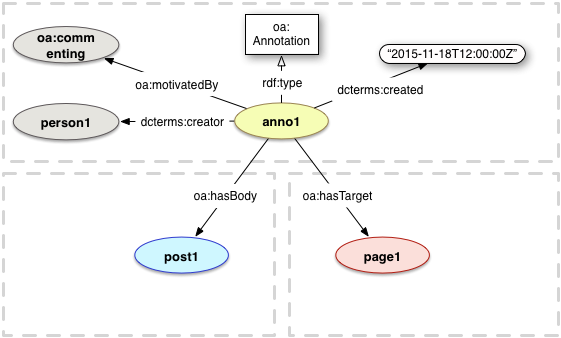
PROV Model Primer
W3C Working Group Note 30 April 2013

Who is using Research Objects?












Workshop on Research Objects (RO2018)
2018-10-29
IEEE eScience Conference 2018,
Amsterdam, Netherlands



Prospective provenance
Capture the method
Yong Zhao, Michael Wilde, Ian Foster (2006):
Applying the virtual data provenance model.
International Provenance and Annotation Workshop (IPAW) 2006

cwlVersion: v1.0
class: Workflow
inputs:
inp: File
ex: string
outputs:
classout:
type: File
outputSource: compile/classfile
steps:
untar:
run: tar-param.cwl
in:
tarfile: inp
extractfile: ex
out: [example_out]
compile:
run: arguments.cwl
in:
src: untar/example_out
out: [classfile]









{
"@context" : [ "https://w3id.org/bundle/context" ],
"id" : "/",
"manifest" : [ "manifest.json" ],
"createdOn" : "2017-08-24T10:57:46.325Z",
"createdBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"authoredBy" : [ {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
}, {
"uri" : "mailto:luka.stojanovic@sbgenomics.com",
"name" : "Luka Stojanovic"
}, {
"uri" : "mailto:crusoe@ucdavis.edu",
"name" : "Michael R. Crusoe"
}, {
"uri" : "mailto:porter@porter.st",
"name" : "Andrey Kartashov"
}, {
"uri" : "mailto:janko.simonovic@sbgenomics.com",
"name" : "Janko Simonovic"
} ],
"retrievedFrom" : "https://github.com/common-workflow-language/workflows/blob/lobstr-v1/workflows/lobSTR/",
"retrievedOn" : "2017-08-24T10:57:46.325Z",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"history" : [ "http:/git2prov.org/git2prov?giturl=https:/github.com/common-workflow-language/workflows.git&serialization=PROV-JSON" ],
"aggregates" : [ {
"uri" : "/workflow/tmp_2.fq",
"mediatype" : "application/octet-stream",
"createdOn" : "2017-08-24T10:57:46.923Z",
"authoredBy" : [ {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/tmp_2.fq",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"bundledAs" : {
"uri" : "urn:uuid:61579f3e-63e6-49c2-b780-f67b2df461b7",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/lobSTR-demo.json",
"mediatype" : "application/json",
"createdOn" : "2017-08-24T10:57:47.216Z",
"authoredBy" : [ {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/lobSTR-demo.json",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"bundledAs" : {
"uri" : "urn:uuid:973caa0e-f3bd-45e8-8d29-70123bc8715a",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/models/illumina_v3.pcrfree.stuttermodel",
"mediatype" : "application/octet-stream",
"createdOn" : "2017-08-24T10:57:47.239Z",
"authoredBy" : [ {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/models/illumina_v3.pcrfree.stuttermodel",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"bundledAs" : {
"uri" : "urn:uuid:62bbcbea-f34f-463f-990d-6148f8ed5e5c",
"folder" : "/workflow/models/"
}
}, {
"uri" : "/workflow/models/illumina_v3.pcrfree.stepmodel",
"mediatype" : "application/octet-stream",
"createdOn" : "2017-08-24T10:57:47.266Z",
"authoredBy" : [ {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/models/illumina_v3.pcrfree.stepmodel",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"bundledAs" : {
"uri" : "urn:uuid:03439ae7-cd94-42a3-b5fe-40bfff6882d8",
"folder" : "/workflow/models/"
}
}, {
"uri" : "/workflow/samtools-sort.cwl",
"mediatype" : "text/x-yaml",
"createdOn" : "2017-08-24T10:57:47.269Z",
"authoredBy" : [ {
"uri" : "mailto:luka.stojanovic@sbgenomics.com",
"name" : "Luka Stojanovic"
}, {
"uri" : "mailto:crusoe@ucdavis.edu",
"name" : "Michael R. Crusoe"
}, {
"uri" : "mailto:porter@porter.st",
"name" : "Andrey Kartashov"
}, {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/samtools-sort.cwl",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"conformsTo" : "https://w3id.org/cwl/v1.0",
"bundledAs" : {
"uri" : "urn:uuid:2dc07859-efc2-4945-a95f-ba7815b68d07",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/lobSTR-workflow.cwl",
"mediatype" : "text/x-yaml",
"createdOn" : "2017-08-24T10:57:47.42Z",
"authoredBy" : [ {
"uri" : "mailto:luka.stojanovic@sbgenomics.com",
"name" : "Luka Stojanovic"
}, {
"uri" : "mailto:crusoe@ucdavis.edu",
"name" : "Michael R. Crusoe"
}, {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/lobSTR-workflow.cwl",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"conformsTo" : "https://w3id.org/cwl/v1.0",
"bundledAs" : {
"uri" : "urn:uuid:58bc1895-3460-46d6-91d7-fa1718d09631",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/lobSTR-arvados-demo.json",
"mediatype" : "application/json",
"createdOn" : "2017-08-24T10:57:47.453Z",
"authoredBy" : [ {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/lobSTR-arvados-demo.json",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"bundledAs" : {
"uri" : "urn:uuid:30c683bc-69fb-4d93-8dad-65b663783af5",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/samtools-index.cwl",
"mediatype" : "text/x-yaml",
"createdOn" : "2017-08-24T10:57:47.458Z",
"authoredBy" : [ {
"uri" : "mailto:luka.stojanovic@sbgenomics.com",
"name" : "Luka Stojanovic"
}, {
"uri" : "mailto:crusoe@ucdavis.edu",
"name" : "Michael R. Crusoe"
}, {
"uri" : "mailto:porter@porter.st",
"name" : "Andrey Kartashov"
}, {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/samtools-index.cwl",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"conformsTo" : "https://w3id.org/cwl/v1.0",
"bundledAs" : {
"uri" : "urn:uuid:8235d3f8-6927-4f73-b160-8521838a1cbb",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/lobSTR-tool.cwl",
"mediatype" : "text/x-yaml",
"createdOn" : "2017-08-24T10:57:47.476Z",
"authoredBy" : [ {
"uri" : "mailto:luka.stojanovic@sbgenomics.com",
"name" : "Luka Stojanovic"
}, {
"uri" : "mailto:crusoe@ucdavis.edu",
"name" : "Michael R. Crusoe"
}, {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/lobSTR-tool.cwl",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"conformsTo" : "https://w3id.org/cwl/v1.0",
"bundledAs" : {
"uri" : "urn:uuid:7fa6fbe4-1fc5-4cb5-9c1a-56b96c5f7aaf",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/allelotype.cwl",
"mediatype" : "text/x-yaml",
"createdOn" : "2017-08-24T10:57:47.537Z",
"authoredBy" : [ {
"uri" : "mailto:luka.stojanovic@sbgenomics.com",
"name" : "Luka Stojanovic"
}, {
"uri" : "mailto:janko.simonovic@sbgenomics.com",
"name" : "Janko Simonovic"
}, {
"uri" : "mailto:crusoe@ucdavis.edu",
"name" : "Michael R. Crusoe"
}, {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/allelotype.cwl",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"conformsTo" : "https://w3id.org/cwl/v1.0",
"bundledAs" : {
"uri" : "urn:uuid:3706bd2f-e53f-431d-b32a-deb661d9b292",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/README",
"mediatype" : "application/octet-stream",
"createdOn" : "2017-08-24T10:57:47.555Z",
"authoredBy" : [ {
"uri" : "mailto:crusoe@ucdavis.edu",
"name" : "Michael R. Crusoe"
}, {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/README",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"bundledAs" : {
"uri" : "urn:uuid:ed54c4d6-c585-4dc9-b7bc-0cf299e20b91",
"folder" : "/workflow/"
}
}, {
"uri" : "/workflow/tmp_1.fq",
"mediatype" : "application/octet-stream",
"createdOn" : "2017-08-24T10:57:47.738Z",
"authoredBy" : [ {
"uri" : "mailto:peter.amstutz@curoverse.com",
"name" : "Peter Amstutz"
} ],
"retrievedFrom" : "https://raw.githubusercontent.com/common-workflow-language/workflows/lobstr-v1/workflows/lobSTR/tmp_1.fq",
"retrievedBy" : {
"uri" : "https://view.commonwl.org",
"name" : "Common Workflow Language Viewer"
},
"bundledAs" : {
"uri" : "urn:uuid:5d431f81-ad0b-4acf-903a-9d5aa03b04df",
"folder" : "/workflow/"
}
}, {
"uri" : "/visualisation.png",
"mediatype" : "image/png",
"createdOn" : "2017-08-24T10:57:47.801Z",
"retrievedFrom" : "https://view.commonwl.org/graph/png/github.com/common-workflow-language/workflows/blob/lobstr-v1/workflows/lobSTR/lobSTR-workflow.cwl",
"bundledAs" : {
"uri" : "urn:uuid:ff9ace37-e76c-49f8-8d36-60f11ff6d257",
"folder" : "/"
}
}, {
"uri" : "/visualisation.svg",
"mediatype" : "image/svg+xml",
"createdOn" : "2017-08-24T10:57:47.821Z",
"retrievedFrom" : "https://view.commonwl.org/graph/svg/github.com/common-workflow-language/workflows/blob/lobstr-v1/workflows/lobSTR/lobSTR-workflow.cwl",
"bundledAs" : {
"uri" : "urn:uuid:a6cfb437-8818-4ab2-9081-efc74c5109e8",
"folder" : "/"
}
} ],
"annotations" : [ {
"uri" : "urn:uuid:9f602fff-b280-41c5-9590-ab95a49c85ad",
"about" : "/",
"content" : "annotations/merged.cwl"
}, {
"uri" : "urn:uuid:0ce4b727-ff61-4534-9afb-e3d676d2782d",
"about" : "/",
"content" : "annotations/workflow.ttl"
} ]
}

#!/usr/bin/env cwl-runner
cwlVersion: v1.0
class: Workflow
label: "Hello World"
doc: "Outputs a message using echo"
inputs: []
outputs:
response:
outputSource: step0/response
type: File
steps:
step0:
run:
class: CommandLineTool
inputs:
message:
type: string
doc: "The message to print"
default: "Hello World"
inputBinding:
position: 1
baseCommand: echo
stdout: response.txt
outputs:
response:
type: stdout
in: []
out: [response]



Retrospective provenance
Capture what happened

Farah Z Khan


Identifying intermediate data
Output 1B file is also Input 2C and Input 3D downstream
Simple filenames -> duplications
./data/step1/outputB.txt
./data/step2/inputC.txt
./data/step3/inputD.txt
Content-adressable
SHA-256 hash of bytes as filename:
./data/51/51fb8af0c4ae0422fbe88340d91880ecb9d7537cf57339c1cf1256b7ca58f32d
RFC6920 URI as global identifier:
nih:sha-256;51fb8af0c4ae0422fbe88340d91880ecb9d7537cf57339c1cf1256b7ca58f32d
arcp://uuid,32a423d6-52ab-47e3-a9cd-54f418a48571/biocompute.json
arcp://uuid,32a423d6-52ab-47e3-a9cd-54f418a48571/data/sequence.bamarcp://ni,sha-256;f4OxZX_x_FO5LcGBSKHWXfwtSx-j1ncoSt3SABJtkGk/biocompute.json
arcp://ni,sha-256;f4OxZX_x_FO5LcGBSKHWXfwtSx-j1ncoSt3SABJtkGk/data/sequence.jsonstain@biggie:~$ sha256sum biocompute-archive.zip
7f83b1657ff1fc53b92dc18148a1d65dfc2d4b1fa3d677284addd200126d9069

cwlVersion: v1.0
class: Workflow
label: EMG QC workflow, (paired end version). Benchmarking with MG-RAST expt.
requirements:
- class: SubworkflowFeatureRequirement
- class: SchemaDefRequirement
types:
- $import: ../tools/FragGeneScan-model.yaml
- $import: ../tools/trimmomatic-sliding_window.yaml
- $import: ../tools/trimmomatic-end_mode.yaml
- $import: ../tools/trimmomatic-phred.yaml
inputs:
reads:
type: File
format: edam:format_1930 # FASTQ
outputs:
processed_sequences:
type: File
outputSource: clean_fasta_headers/sequences_with_cleaned_headers
steps:
trim_quality_control:
doc: |
Low quality trimming (low quality ends and sequences with < quality scores
less than 15 over a 4 nucleotide wide window are removed)
run: ../tools/trimmomatic.cwl
in:
reads1: reads
phred: { default: '33' }
leading: { default: 3 }
trailing: { default: 3 }
end_mode: { default: SE }
minlen: { default: 100 }
slidingwindow:
default:
windowSize: 4
requiredQuality: 15
out: [reads1_trimmed]
convert_trimmed-reads_to_fasta:
run: ../tools/fastq_to_fasta.cwl
in:
fastq: trim_quality_control/reads1_trimmed
out: [ fasta ]
clean_fasta_headers:
run: ../tools/clean_fasta_headers.cwl
in:
sequences: convert_trimmed-reads_to_fasta/fasta
out: [ sequences_with_cleaned_headers ]
$namespaces:
edam: http://edamontology.org/
s: http://schema.org/
$schemas:
- http://edamontology.org/EDAM_1.16.owl
- https://schema.org/docs/schema_org_rdfa.html
s:license: "https://www.apache.org/licenses/LICENSE-2.0"
s:copyrightHolder: "EMBL - European Bioinformatics Institute"


{
"id": "https://w3id.org/cwl/view/git/886df9de6713e06228d2560c40f451155a196383/workflows/emg-qc-single.cwl",
"name": "EMG QC workflow, (paired end version)",
"structured_name": "EMG QC workflow, (paired end version). Benchmarking with MG-RAST expt.",
"version": "0.0.886df9de6713e06228d2560c40f451155a196383",
"digital_signature": "886df9de6713e06228d2560c40f451155a196383",
"verification_status": "unreviewed",
"publicaton_status": "open_access",
"authors": ["https://orcid.org/0000-0001-8626-2148",
"https://orcid.org/0000-0002-2961-9670"],
"usability_domain": ["metagenomics"],
"description_domain": {
"keywords": ["quality control", "trimming"],
"github_extension": {
"github_repository": "EBI-Metagenomics/ebi-metagenomics-cwl",
"github_url": "https://github.com/EBI-Metagenomics/ebi-metagenomics-cwl/",
"github_uri": ["workflows/emg-qc-single.cwl"]
},
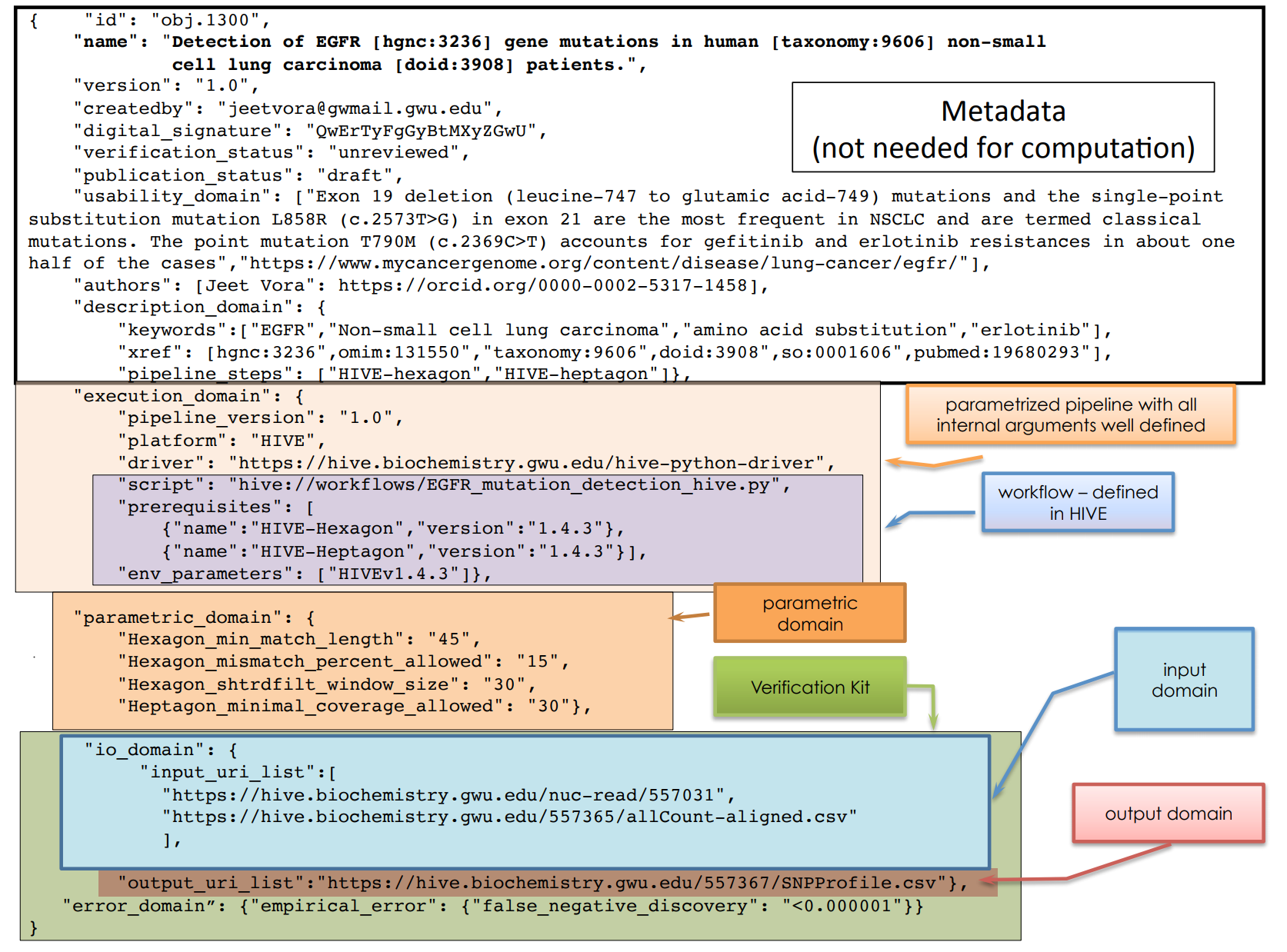
}Workflow metadata as BioCompute Object

"pipeline_steps": [
{ "tool_name": "trim_quality_control",
"tool_desc": "Low quality trimming (low quality ends and sequences with < quality scores
less than 15 over a 4 nucleotide wide window are removed)",
"tool_version": "0.32",
"tool_package": ["trimmomatic [rrid:RRID:SCR_011848] version 0.32+dfsg-1",
"openjdk-7-jre-headless"],
"step_number": "1",
"input_uri": {
"reference_files": [],
"input_file_list": [
"arcp://21f3a974-7b58-58d1-9687-7d5735aad6bd/tools/otu_table.biom"
]
},
"output_uri_list": [
"arcp://97b36186-13d9-497c-b2bb-7d41c995f4ee/data/51/51fb8af0c4ae0422fbe88340d91880ecb9d7537cf57339c1cf1256b7ca58f32d"]
},
{ "tool_name": "convert_trimmed-reads_to_fasta"
"input_uri": {
"input_file_list": [
"arcp://97b36186-13d9-497c-b2bb-7d41c995f4ee/data/51/51fb8af0c4ae0422fbe88340d91880ecb9d7537cf57339c1cf1256b7ca58f32d"]
}, ..
},
{ "tool_name": "clean_fasta_headers"
..
},
],Prospective(?) provenance as BioCompute Object

"execution_domain": {
"script_type": "URI",
"script": "https://raw.githubusercontent.com/EBI-Metagenomics/ebi-metagenomics-cwl/c34db66a79cec3b66a0f1be5e499eef88db5a9ed/workflows/emg-qc-single.cwl",
"script": "https://w3id.org/cwl/view/git/886df9de6713e06228d2560c40f451155a196383/workflows/emg-qc-single.cwl?format=yaml",
"platform": "cwl",
"driver": "cwl",
"software_prerequisities": [
{"name": "trimmomatic", "version":"0.32"},
{"name": "openjdk-7-jre-headless", "version": "7"},
{"name": "biopython", "version": "1.69"}
],
"env_parameters": [
{
"ramMin": "10240",
"coresMin": "8"
}
]
}Execution domain for CWL workflow
"parametric_domain": {
"phred": "33",
"leading": "3",
"trailing": "3",
"end_mode": "SE",
"minlen": "100",
"slidingwindow": {
"windowSize": "4",
"requiredQuality": "15"
}
}Parametric domain for CWL workflow
inputs:
phred:
type: trimmomatic-phred.yaml#phred?
doc: "33" or "64" specifies the base quality encoding. Default: 64
leading:
type: int?
doc: |
Remove low quality bases from the beginning. As long as a base has a value
below this threshold the base is removed and the next base will be investigated.
trailing:
type: int?
doc: |
Remove low quality bases from the end. As long as a base has a value
below this threshold the base is removed and the next base (which as
trimmomatic is starting from the 3' prime end would be base preceding
the just removed base) will be investigated. This approach can be used
removing the special Illumina "low quality segment" regions (which are
marked with quality score of 2), but we recommend Sliding Window or
MaxInfo instead
end_mode:
type: trimmomatic-end_mode.yaml#end_mode
doc: |
Single End (SE) or Paired End (PE) mode
minlen:
type: int?
doc: |
This module removes reads that fall below the specified minimal length.
If required, it should normally be after all other processing steps.
Reads removed by this step will be counted and included in the "dropped
reads" count presented in the trimmomatic summary.
slidingwindow:
type: trimmomatic-sliding_window.yaml#slidingWindow?
doc: |
Perform a sliding window trimming, cutting once the average quality
within the window falls below a threshold. By considering multiple
bases, a single poor quality base will not cause the removal of high
quality data later in the read.
<windowSize> specifies the number of bases to average across
<requiredQuality> specifies the average quality required

"io_domain": {
"input_subdomain": {
"reference_files": [],
"input_file_list": {
"reads": [
"arcp://21f3a974-7b58-58d1-9687-7d5735aad6bd/tools/otu_table.biom"
]
}
},
"output_subdomain": {
"processed_sequences": [
{
"title": "sequences_with_cleaned_headers [edam:format_1929]"
"uri": "arcp://97b36186-13d9-497c-b2bb-7d41c995f4ee/data/7c/7cf57339c1cf1256b7ca58f32d51fb8af0c4ae0422fbe88340d91880ecb9d753"
"mime-type": "text/x-fasta"
},
],
}IO domain for CWL workflow

13:15 -> 14:00
Breakout group D
Leverage existing efforts to realize the BioCompute vision
What existing standards, efforts, and specifications can contribute to the BCO vision?



