Stian Soiland-Reyes
eScience lab, The University of Manchester
BioExcel/MolSSI symposium, PASC18
2018-07-03 Basel, CH
This work is licensed under a
Creative Commons Attribution 4.0 International License.



This work has been done as part of the BioExcel CoE (www.bioexcel.eu), a project funded by the European Union contract H2020-EINFRA-2015-1-675728.

Facing Compute Platform Portability Challenges with Scientific Workflows
Experiences from Common Workflow Language


cwlVersion: v1.0
class: Workflow
inputs:
inp: File
ex: string
outputs:
classout:
type: File
outputSource: compile/classfile
steps:
untar:
run: tar-param.cwl
in:
tarfile: inp
extractfile: ex
out: [example_out]
compile:
run: arguments.cwl
in:
src: untar/example_out
out: [classfile]

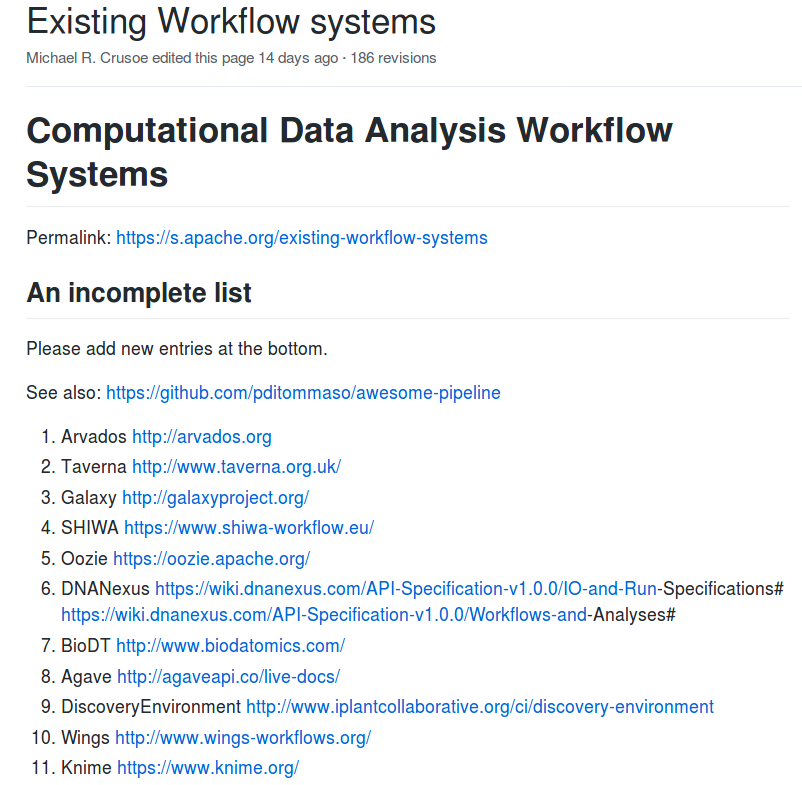
cwltool: Local (Linux, OS X, Windows)
Arvados: AWS, GCP, Azure, Slurm
Toil: AWS, Azure, GCP, Grid Engine, LSF, Mesos, OpenStack, Slurm, PBS/Torque, HTcondor
Rabix Bunny: Local(Linux, OS X), GA4GH TES
cwl-tes: Local, GCP, AWS, HTCondor, Grid Engine, PBS/Torque, Slurm
CWL-Airflow: Linux, OS X
REANA: Kubernetes, CERN OpenStack
cromwell: local, HPC, Google, HtCondor
CWLEXEC: IBM Spectrum LSF
XENON: any Xenon backend: local, ssh, SLURM, Torque, Grid Engine
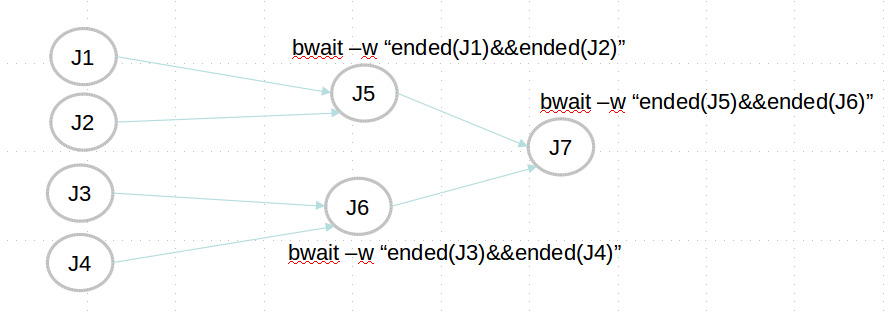
Which CWL engine runs where?







☑ Efficient checking of job completion with maximum parallelism
☑ Support LSF submission (bsub) options
☑ Self-healing of workflows
☑ Docker integration
☑ Cloud bursting
☑ Rerun and interruption
cwlexec
{
"queue": "high",
"steps": {
"step1": {
"app": "dockerapp"
},
"step2": {
"res_req": "select[type==X86_64] order[ut]
rusage[mem=512MB:swp=1GB:tmp=500GB]"
}
}
}
cwlexec Run Profile
#!/usr/bin/env cwl-runner
cwlVersion: v1.0
class: Workflow
inputs:
inp: File
ex: string
outputs:
classout:
type: File
outputSource: compile/classfile
steps:
untar:
run: tar-param.cwl
in:
tarfile: inp
extractfile: ex
out: [example_out]
compile:
run: arguments.cwl
in:
src: untar/example_out
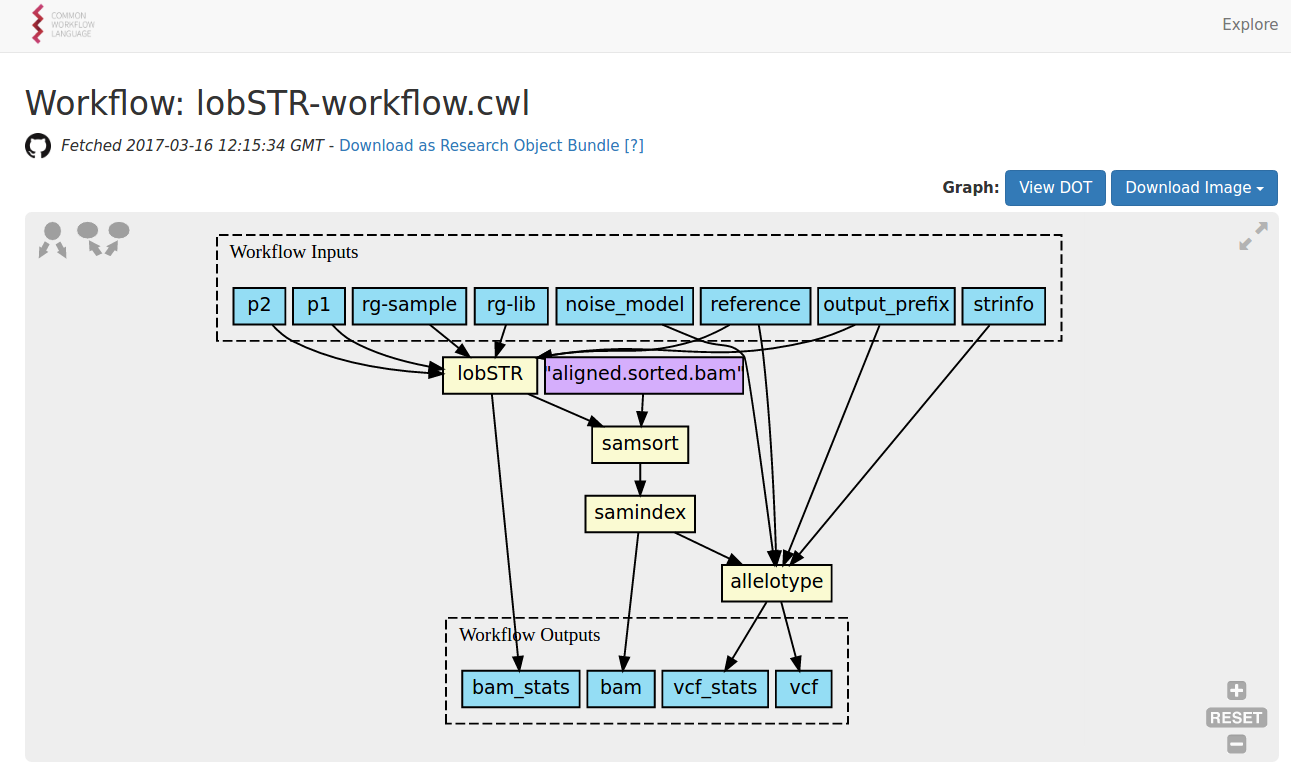
out: [classfile]Composing a workflow
cwlVersion: v1.0
class: Workflow
label: EMG QC workflow, (paired end version). Benchmarking with MG-RAST expt.
requirements:
- class: SubworkflowFeatureRequirement
- class: SchemaDefRequirement
types:
- $import: ../tools/FragGeneScan-model.yaml
- $import: ../tools/trimmomatic-sliding_window.yaml
- $import: ../tools/trimmomatic-end_mode.yaml
- $import: ../tools/trimmomatic-phred.yaml
inputs:
reads:
type: File
format: edam:format_1930 # FASTQ
outputs:
processed_sequences:
type: File
outputSource: clean_fasta_headers/sequences_with_cleaned_headers
steps:
trim_quality_control:
doc: |
Low quality trimming (low quality ends and sequences with < quality scores
less than 15 over a 4 nucleotide wide window are removed)
run: ../tools/trimmomatic.cwl
in:
reads1: reads
phred: { default: '33' }
leading: { default: 3 }
trailing: { default: 3 }
end_mode: { default: SE }
minlen: { default: 100 }
slidingwindow:
default:
windowSize: 4
requiredQuality: 15
out: [reads1_trimmed]
convert_trimmed-reads_to_fasta:
run: ../tools/fastq_to_fasta.cwl
in:
fastq: trim_quality_control/reads1_trimmed
out: [ fasta ]
clean_fasta_headers:
run: ../tools/clean_fasta_headers.cwl
in:
sequences: convert_trimmed-reads_to_fasta/fasta
out: [ sequences_with_cleaned_headers ]
$namespaces:
edam: http://edamontology.org/
s: http://schema.org/
$schemas:
- http://edamontology.org/EDAM_1.16.owl
- https://schema.org/docs/schema_org_rdfa.html
s:license: "https://www.apache.org/licenses/LICENSE-2.0"
s:copyrightHolder: "EMBL - European Bioinformatics Institute"

#!/usr/bin/env cwl-runner
cwlVersion: v1.0
class: CommandLineTool
baseCommand: [tar, xf]
inputs:
tarfile:
type: File
inputBinding:
position: 1
outputs:
example_out:
type: File
outputBinding:
glob: hello.txtCommand line tool
class: CommandLineTool
hints:
SoftwareRequirement:
packages:
samtools:
version: [ "0.1.19" ]
baseCommand: ["samtools", "index"]
#..Finding the tool
module load samtools/0.1.19apt-get install samtools=0.1.19*conda install samtools=0.1.19<dependency_resolvers>
<modules modulecmd="/opt/bin/modulecmd" />
<tool_shed_packages />
<galaxy_packages />
<conda />
<modules modulecmd="/opt/bin/modulecmd" versionless="true" />
<galaxy_packages versionless="true" />
<conda versionless="true" />
</dependency_resolvers>Package resolution
tool_dependency_dir/
samtools/
0.1.19/
bin/
env.sh
Dependency resolution by CWLTool and Toil



Where to find command line tools?

cwlVersion: v1.0
class: CommandLineTool
baseCommand: node
hints:
DockerRequirement:
dockerPull: mgibio/samtools:1.3.1



= anaconda

Let's add some identifiers!
hints:
SoftwareRequirement:
packages:
- package: bowtie
version:
- '2.2.8'
specs:
- https://packages.debian.org/bowtie
- https://anaconda.org/bioconda/bowtie
- https://bio.tools/tool/bowtie2/version/2.2.8
- https://identifiers.org/rrid/RRID:SCR_005476
- https://hpc.example.edu/modules/bowtie-tbb/2.2
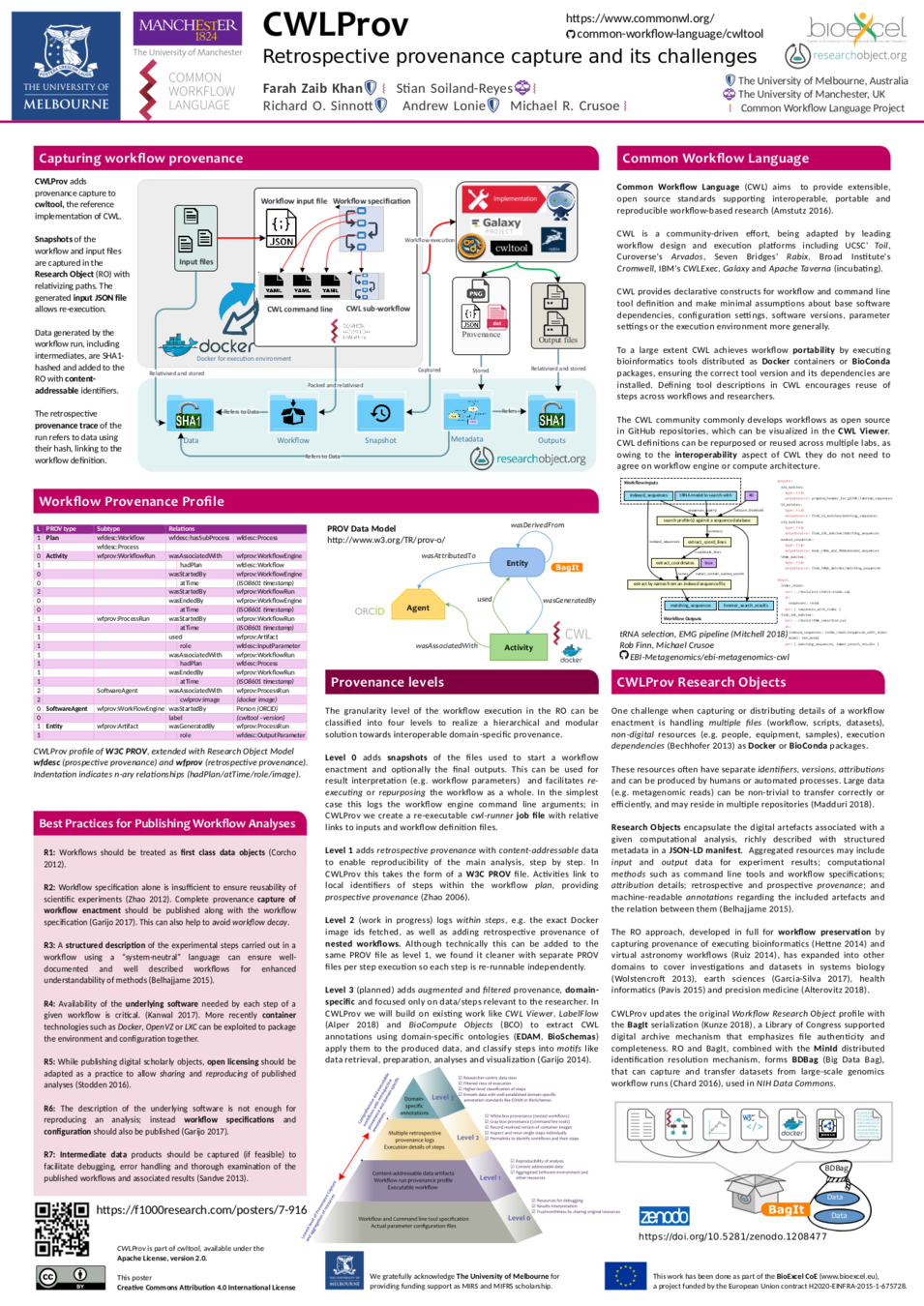
Khan et al,
CWLProv – Interoperable retrospective provenance capture and its challenges,
BOSC 2018

document
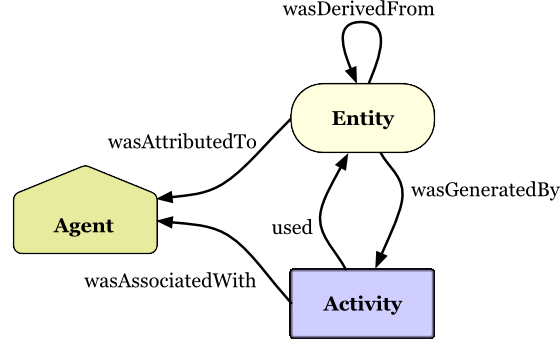
prefix wfprov <http://purl.org/wf4ever/wfprov#>
prefix prov <http://www.w3.org/ns/prov#>
prefix wfdesc <http://purl.org/wf4ever/wfdesc#>
prefix wf <https://w3id.org/cwl/view/git/933bf2a1a1cce32d88f88f136275535da9df0954/workflows/hello/hello.cwl#>
prefix input <app://579c1b74-b328-4da6-80a8-a2ffef2ac9b5/workflow/input.json#>
prefix run <urn:uuid:>
prefix engine <urn:uuid:>
prefix data <urn:hash:sha256:>
default <app://579c1b74-b328-4da6-80a8-a2ffef2ac9b5/>
// Level 1 provenance of workflow run
activity(run:2e1287e0-6dfb-11e7-8acf-0242ac110002, , , [prov:type='wfprov:WorkflowRun', prov:label="Run of workflow/packed.cwl#main"])
wasStartedBy(run:2e1287e0-6dfb-11e7-8acf-0242ac110002, -, -, -, 2017-10-27T14:24:00+01:00)
// The engine is the SoftwareAgent that is executing our Workflow plan
wasAssociatedWith(run:2e1287e0-6dfb-11e7-8acf-0242ac110002, engine:b2210211-8acb-4d58-bd28-2a36b18d3b4f, wf:main)
agent(engine:b2210211-8acb-4d58-bd28-2a36b18d3b4f, prov:type='prov:SoftwareAgent', prov:type='wfprov:WorkflowEngine', prov:label="cwltool v1.2.5")
// prov has no term to relate sub-plans - we'll use wfdesc:hasSubProcess
entity(wf:main,[prov:type='wfdesc:Workflow', prov:type='prov:Plan', wfdesc:hasSubProcess='wf:main/step1', wfdesc:hasSubProcess='wf:main/step2'])
alternateOf(wf:main, workflow/packed.cwl)
entity(wf:main/step1,[prov:type='wfdesc:Process', prov:type='prov:Plan'])
entity(wf:main/step2,[prov:type='wfdesc:Process', prov:type='prov:Plan'])
// First the workflow uses some data; here with a urn:sha:sha256 identifier
used(run:2e1287e0-6dfb-11e7-8acf-0242ac110002, data:5891b5b522d5df086d0ff0b110fbd9d21bb4fc7163af34d08286a2e846f6be03, 2017-10-27T14:29:00+01:00, [prov:role='wf:main/input1']))
entity(data:5891b5b522d5df086d0ff0b110fbd9d21bb4fc7163af34d08286a2e846f6be03, [prov:type='wfprov:Artifact'])
// which we have stored a copy of within the research object
specializationOf(data/58/5891b5b522d5df086d0ff0b110fbd9d21bb4fc7163af34d08286a2e846f6be03, data:5891b5b522d5df086d0ff0b110fbd9d21bb4fc7163af34d08286a2e846f6be03)
// Then there was another activity - wfprov:ProcessRun indicating a command line tool
activity(run:4305467e-6dfb-11e7-885d-0242ac110002, -, -, [prov:type='wfprov:ProcessRun', prov:label="Run of workflow/packed.cwl#main/step1"])
// started by the mother activity
wasStartedBy(run:4305467e-6dfb-11e7-885d-0242ac110002, -, -, run:2e1287e0-6dfb-11e7-8acf-0242ac110002, 2017-10-27T15:00:00+01:00)
// same engine using step1 as plan. In a distributed scenario there might be a different engine
wasAssociatedWith(run:4305467e-6dfb-11e7-885d-0242ac110002, engine:b2210211-8acb-4d58-bd28-2a36b18d3b4f, wf:main/step1)
// This activity also use the same data, but in a different role (e.g. input parameter)
used(run:4305467e-6dfb-11e7-885d-0242ac110002, data:5891b5b522d5df086d0ff0b110fbd9d21bb4fc7163af34d08286a2e846f6be03, 2017-10-27T14:00:00+01:00, [prov:role='wf:main/step1/in1'])
// And we generate some new data
wasGeneratedBy(data:00688350913f2f292943a274b57019d58889eda272370af261c84e78e204743c, run:4305467e-6dfb-11e7-885d-0242ac110002, 2017-10-27T16:00:00+01:00, [prov:role='wf:main/step1/out1']))
entity(data:00688350913f2f292943a274b57019d58889eda272370af261c84e78e204743c, [prov:type='wfprov:Artifact'])
// again stored in the RO
specializationOf(data/00/00688350913f2f292943a274b57019d58889eda272370af261c84e78e204743c, data:00688350913f2f292943a274b57019d58889eda272370af261c84e78e204743c)
// step1 finished
wasEndedBy(run:4305467e-6dfb-11e7-885d-0242ac110002, -, -, run:2e1287e0-6dfb-11e7-8acf-0242ac110002, 2017-10-27T15:30:00+01:00)
// the master workflow then "generate" that same value, but now at a different time and role (the resultA master workflow output)
wasGeneratedBy(data:00688350913f2f292943a274b57019d58889eda272370af261c84e78e204743c, run:2e1287e0-6dfb-11e7-8acf-0242ac110002, 2017-10-27T15:00:00+01:00, [prov:role='wf:main/resultA'])
// next step activity
activity(run:c42dc36e-6dfd-11e7-bc24-0242ac110002, -, - [prov:type='wfprov:ProcessRun', prov:label="Run of workflow/packed.cwl#main/step2"])
wasStartedBy(run:c42dc36e-6dfd-11e7-bc24-0242ac110002, -, -, run:2e1287e0-6dfb-11e7-8acf-0242ac110002, 2017-10-27T16:00:00+01:00)
// associated with step2
wasAssociatedWith(run:c42dc36e-6dfd-11e7-bc24-0242ac110002, engine:b2210211-8acb-4d58-bd28-2a36b18d3b4f, wf:main/step2)
// Uses two data artifacts; one which came from previous step, other as workflow input
used(run:4305467e-6dfb-11e7-885d-0242ac110002, data:5891b5b522d5df086d0ff0b110fbd9d21bb4fc7163af34d08286a2e846f6be03, 2017-10-27T15:00:00+01:00, [prov:role='wf:main/step2/valueA'])
used(run:4305467e-6dfb-11e7-885d-0242ac110002, data:00688350913f2f292943a274b57019d58889eda272370af261c84e78e204743c, 2017-10-27T15:00:00+01:00, [prov:role='wf:main/step2/valueB'])
// and generate two new data artifacts
wasGeneratedBy(data:952f537d1f3116db56703787ace248fe00ae46fa77ea3803aa3d8dc01d221a9d, run:c42dc36e-6dfd-11e7-bc24-0242ac110002, 2017-10-27T16:34:20+01:00, [prov:role='wf:main/step2/out1'])))
entity(data:952f537d1f3116db56703787ace248fe00ae46fa77ea3803aa3d8dc01d221a9d, [prov:type='wfprov:Artifact'])
specializationOf(data/95/2f537d1f3116db56703787ace248fe00ae46fa77ea3803aa3d8dc01d221a9d, data:952f537d1f3116db56703787ace248fe00ae46fa77ea3803aa3d8dc01d221a9d)
wasGeneratedBy(data:3deb00bd0decd1f21d015a178c4f23a5eb537588c08eeee9d55059ec29637be0, run:c42dc36e-6dfd-11e7-bc24-0242ac110002, 2017-10-27T16:34:20+01:00, [prov:role='wf:main/step2/out2'])))
entity(data:3deb00bd0decd1f21d015a178c4f23a5eb537588c08eeee9d55059ec29637be0, [prov:type='wfprov:Artifact'])
specializationOf(data/3d/eb00bd0decd1f21d015a178c4f23a5eb537588c08eeee9d55059ec29637be0, data:3deb00bd0decd1f21d015a178c4f23a5eb537588c08eeee9d55059ec29637be0)
// step2 ends
wasEndedBy(run:c42dc36e-6dfd-11e7-bc24-0242ac110002, -, -, run:2e1287e0-6dfb-11e7-8acf-0242ac110002, 2017-10-27T16:30:00+01:00)
// only step output out1 captured by mother workflow, sent to resultB workflow output
wasGeneratedBy(data:952f537d1f3116db56703787ace248fe00ae46fa77ea3803aa3d8dc01d221a9d, run:2e1287e0-6dfb-11e7-8acf-0242ac110002, 2017-10-27T15:00:00+01:00, [prov:role='wf:main/resultB'])
// mother workflow ends
wasEndedBy(run:2e1287e0-6dfb-11e7-8acf-0242ac110002, -, -, run:2e1287e0-6dfb-11e7-8acf-0242ac110002, 2017-10-27T16:34:40+01:00)
endDocument
CWLProv


Using provenance to improve performance

Mondelli et al: BioWorkbench
https://arxiv.org/abs/1801.03915
Challenges
Determining hardware allocations
Automatic rescale+retry
Configuration or Prediction?
Machine learning from provenance
Scheduling scattered jobs
Can't determine total number of jobs until runtime
..but usually early on you can find the number
Accessing the scheduler
How can tasks influence their own allocations?
Lesson learnt from cloud approach:
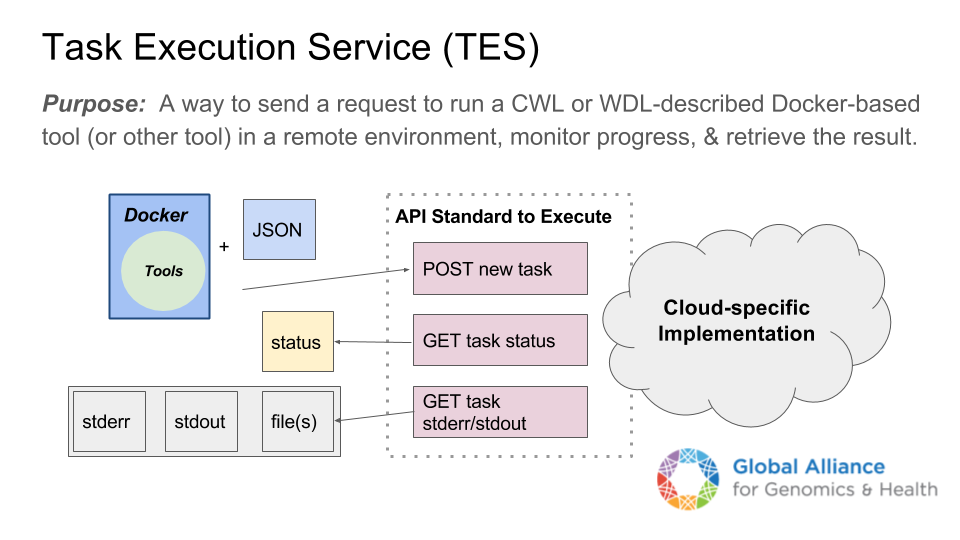
Avoid "master" node dependency
Workarounds: Task Service (TES), worker nodes
Moving workflows to Exascale
Don't throw the baby out with the bathwater!
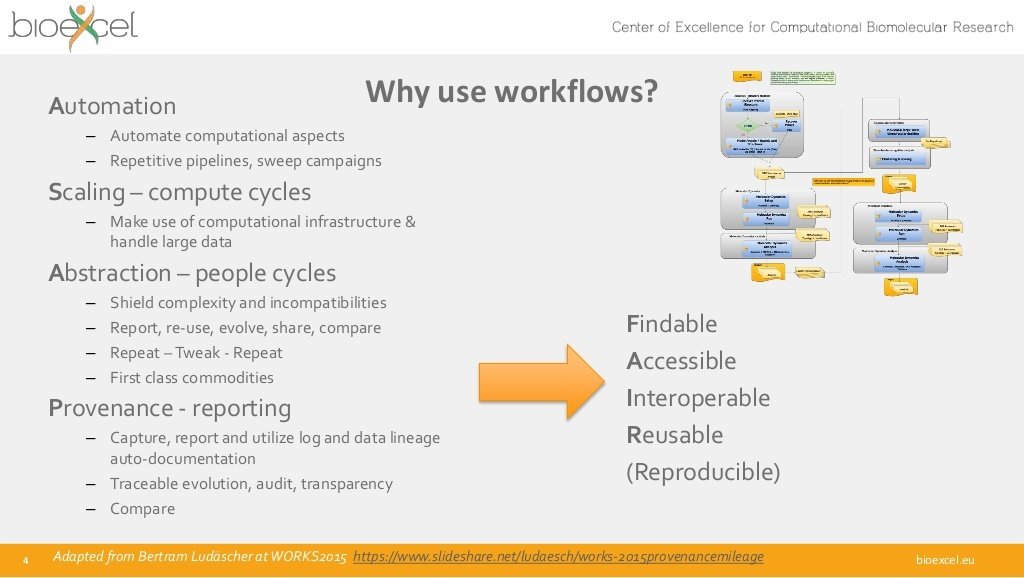
☒ Automation (but is is interoperable?)
☑ Scalability (faster! bigger!)
☒ Abstraction (can humans still understand it?)
☐ Provenance (what actually ran?)
☑ Findable
☒ Accessable
☒ Interoperable
☐ Reproducible