WP7 wrap-up

IBISBA WP6/WP7 workshop, VTT, Espoo
2018-06-15
☑ RetroPath 2.0
☑ RetroPath 2.0-mods-metabolomics
☑ Scope Viewer (with RetroPath)
☑ RP2Path
☑ Selenzyme connector for retropath results
☐ OpenMS
☐ Query Biochemical Rules DataBase (BioRule DB)
~December 2018
Initial Workflow nodes for design








-
Title
-
Description
-
Workflow type (e.g. KNIME, Galaxy)
-
Diagram
-
License (Apache, GPL, CC-BY)
-
License of workflow
-
license of its dependencies
-
Permission needed to use? (e.g. for commercial users)
-
-
Compatibility
-
E.g. Windows or Linux
-
WF environment, e.g. plugins
-
-
Purpose
-
Related to SEEK project structure
-
Workflow metadata


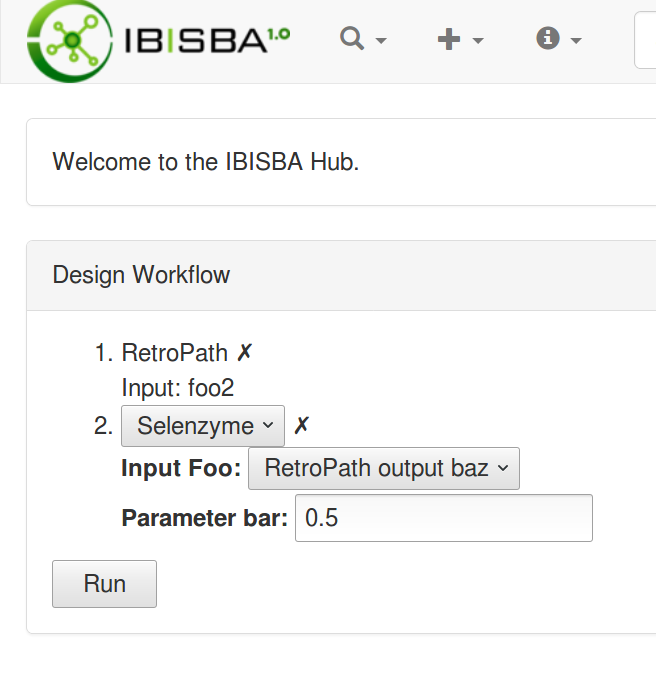
Workflow as chain











Workflow nodes to make
Design
Extract sink from strain models (native metabolites)
Machine learning based Enzyme sequence scoring node
Flux balance analysis node
Pathway ranking node
Build
Connector node with DNA part repository (promoter, enzyme sequence)
From RP2Path/DNA part connector to SBOL
Translate SBOL to/from vector (SEVA)
Generate combinatorial libraries of SBOL/SEVA varying enzymes sequences, ORI, promoter, RBS for bacteria (strain specific)
Format Build and test SOPs in structured language (AutoProtocol?)
...
Test
Untargeted/targeted metabolomics connector RetroPath 2.0-mods-metabolomics with OpenMS
...
Optimize and Scale up
Mixed integer linear program node for gene up/down regulation
DoE for fermentation condition
...
KNIME to CWL

QuickForm inputs
Workflow parameters


Practical: Wrap RP2Path in CWL







Moving from proof of concept to production instance




Single Sign-On for IBISBAHub
Data Storage
by reference




...


Data Management Plan

{
"op": "oligosynthesize",
"oligos": [
{
"destination": "my_plate/A1",
"sequence": "CATGGGTAVNNNNGAAB",
"scale": "25nm" | "100nm" | "250nm" | "1um",
"purification": "standard" | "page" | "hplc",
// default: standard
},
// ...
]
}