Stian Soiland-Reyes
(channelling Carole Goble, Michael Crusoe, Adam Hospital)
eScience lab, The University of Manchester
Pistoia Symposium:
Application of Bioinformatics
in support of Precision Medicine
2019-03-11, London
This work is licensed under a
Creative Commons Attribution 4.0 International License.



This work has been done as part of the BioExcel CoE (www.bioexcel.eu), a project funded by the European Union contracts H2020-INFRAEDI-02-2018-823830, H2020-EINFRA-2015-1-675728

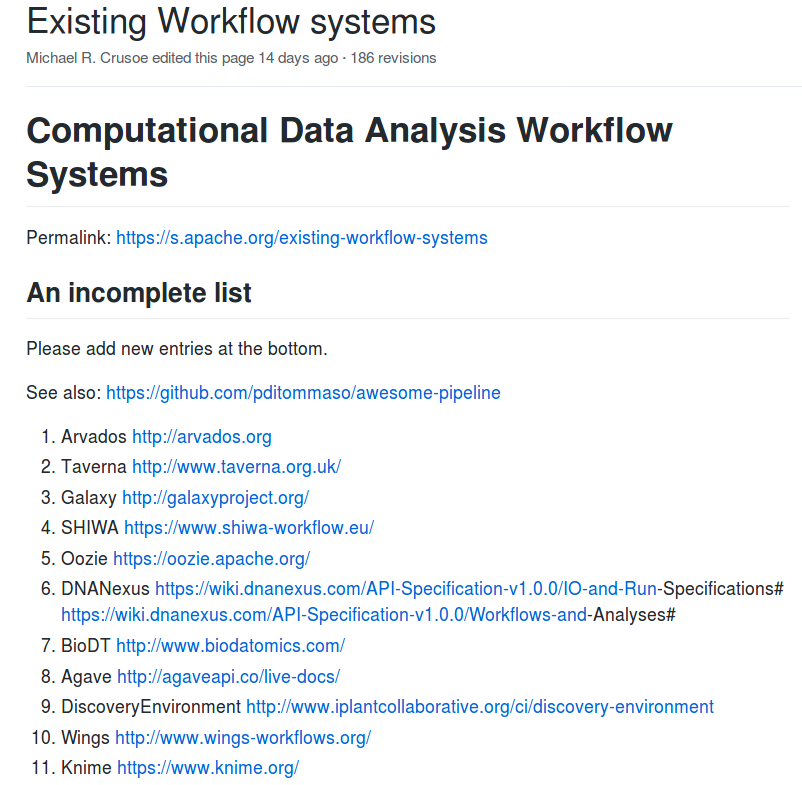
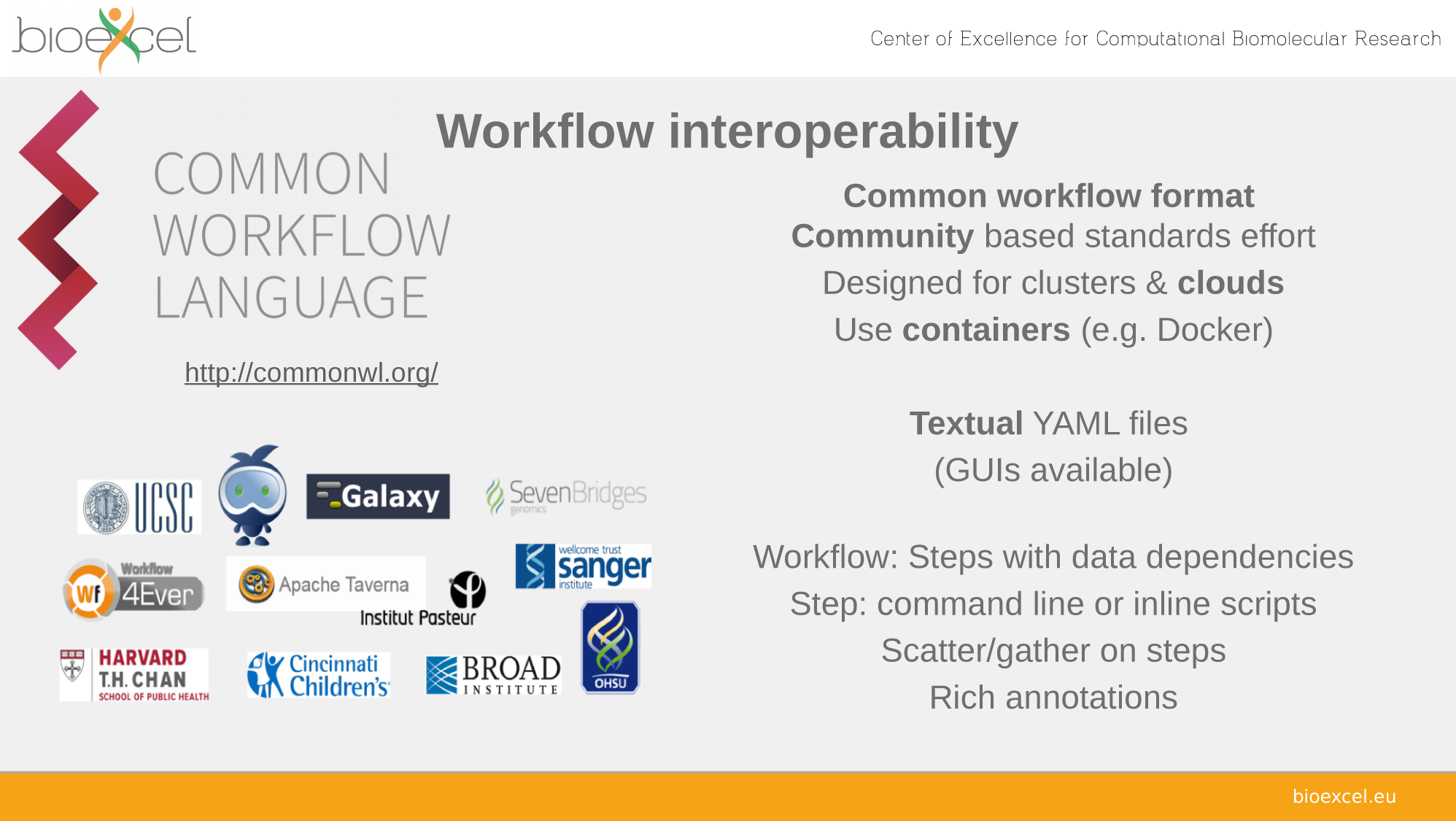
Sharing reproducible computational analyses
☑ Workflows
☑ Containers
☑ Repositories

Workflows
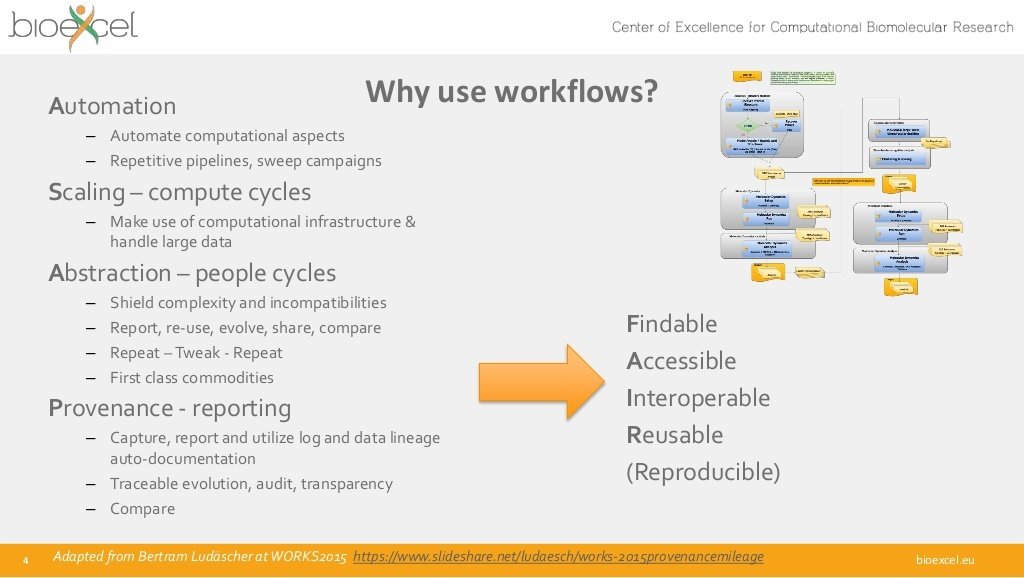

Findable
Accessible
Interoperable
Reusable







cwlVersion: v1.0
class: Workflow
inputs:
inp: File
ex: string
outputs:
classout:
type: File
outputSource: compile/classfile
steps:
untar:
run: tar-param.cwl
in:
tarfile: inp
extractfile: ex
out: [example_out]
compile:
run: arguments.cwl
in:
src: untar/example_out
out: [classfile]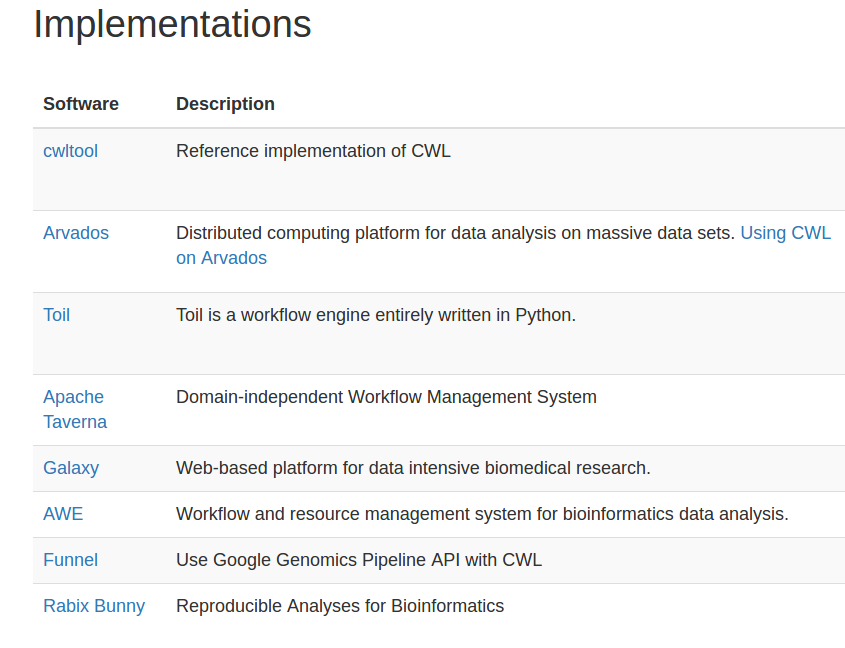


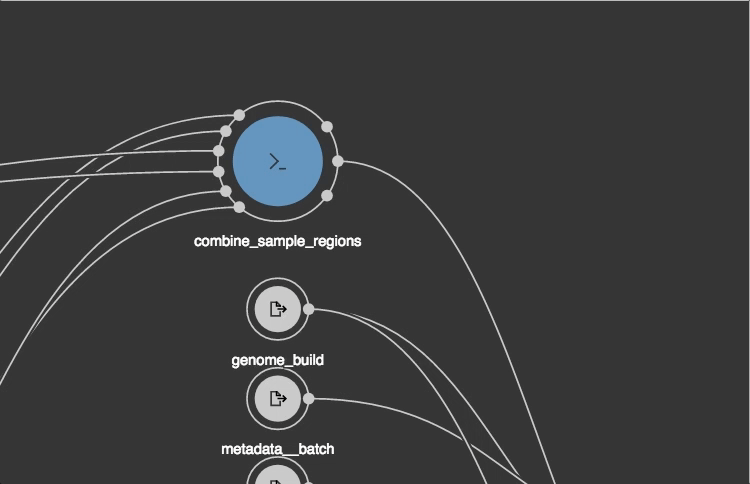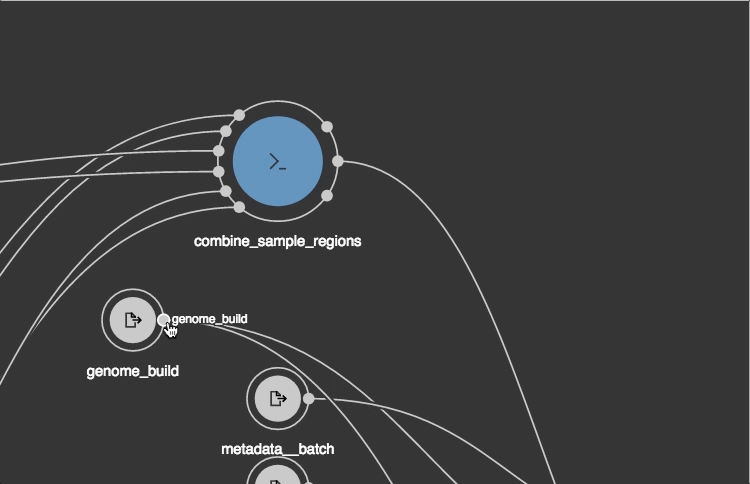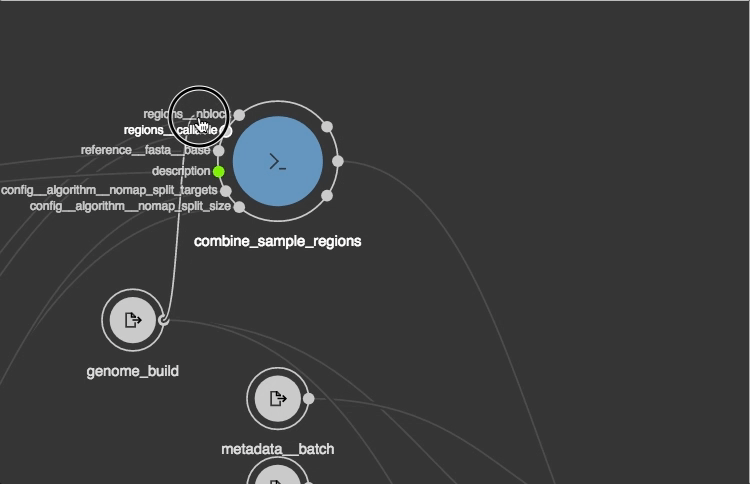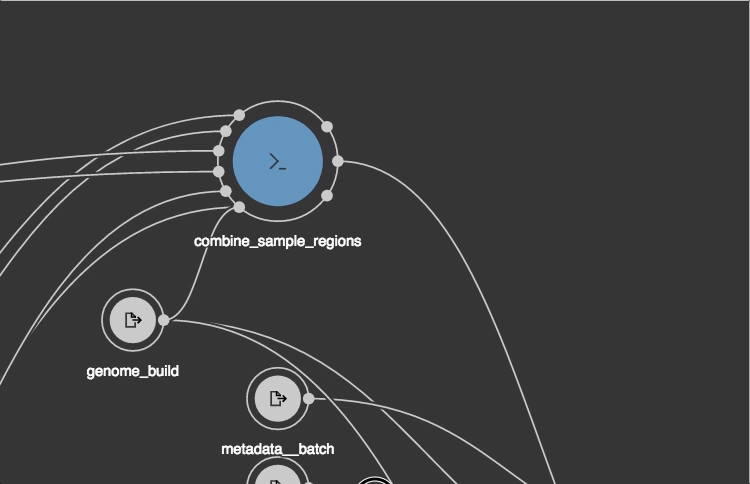

cwlVersion: v1.0
class: Workflow
label: EMG QC workflow, (paired end version). Benchmarking with MG-RAST expt.
requirements:
- class: SubworkflowFeatureRequirement
- class: SchemaDefRequirement
types:
- $import: ../tools/FragGeneScan-model.yaml
- $import: ../tools/trimmomatic-sliding_window.yaml
- $import: ../tools/trimmomatic-end_mode.yaml
- $import: ../tools/trimmomatic-phred.yaml
inputs:
reads:
type: File
format: edam:format_1930 # FASTQ
outputs:
processed_sequences:
type: File
outputSource: clean_fasta_headers/sequences_with_cleaned_headers
steps:
trim_quality_control:
doc: |
Low quality trimming (low quality ends and sequences with < quality scores
less than 15 over a 4 nucleotide wide window are removed)
run: ../tools/trimmomatic.cwl
in:
reads1: reads
phred: { default: '33' }
leading: { default: 3 }
trailing: { default: 3 }
end_mode: { default: SE }
minlen: { default: 100 }
slidingwindow:
default:
windowSize: 4
requiredQuality: 15
out: [reads1_trimmed]
convert_trimmed-reads_to_fasta:
run: ../tools/fastq_to_fasta.cwl
in:
fastq: trim_quality_control/reads1_trimmed
out: [ fasta ]
clean_fasta_headers:
run: ../tools/clean_fasta_headers.cwl
in:
sequences: convert_trimmed-reads_to_fasta/fasta
out: [ sequences_with_cleaned_headers ]
$namespaces:
edam: http://edamontology.org/
s: http://schema.org/
$schemas:
- http://edamontology.org/EDAM_1.16.owl
- https://schema.org/docs/schema_org_rdfa.html
s:license: "https://www.apache.org/licenses/LICENSE-2.0"
s:copyrightHolder: "EMBL - European Bioinformatics Institute"

cwltool: Local (Linux, OS X, Windows)
Arvados: AWS, GCP, Azure, Slurm
Toil: AWS, Azure, GCP, Grid Engine, LSF, Mesos, OpenStack, Slurm, PBS/Torque
Rabix Bunny: Local(Linux, OS X), GA4GH TES
cwl-tes: Local, GCP, AWS, HTCondor, Grid Engine, PBS/Torque, Slurm
CWL-Airflow: Linux, OS X
REANA: Kubernetes, CERN OpenStack
cromwell: local, HPC, Google, HtCondor
CWLEXEC: IBM Spectrum LSF
XENON: any Xenon backend: local, ssh, SLURM, Torque, Grid Engine
Which CWL engine runs where?
Containers

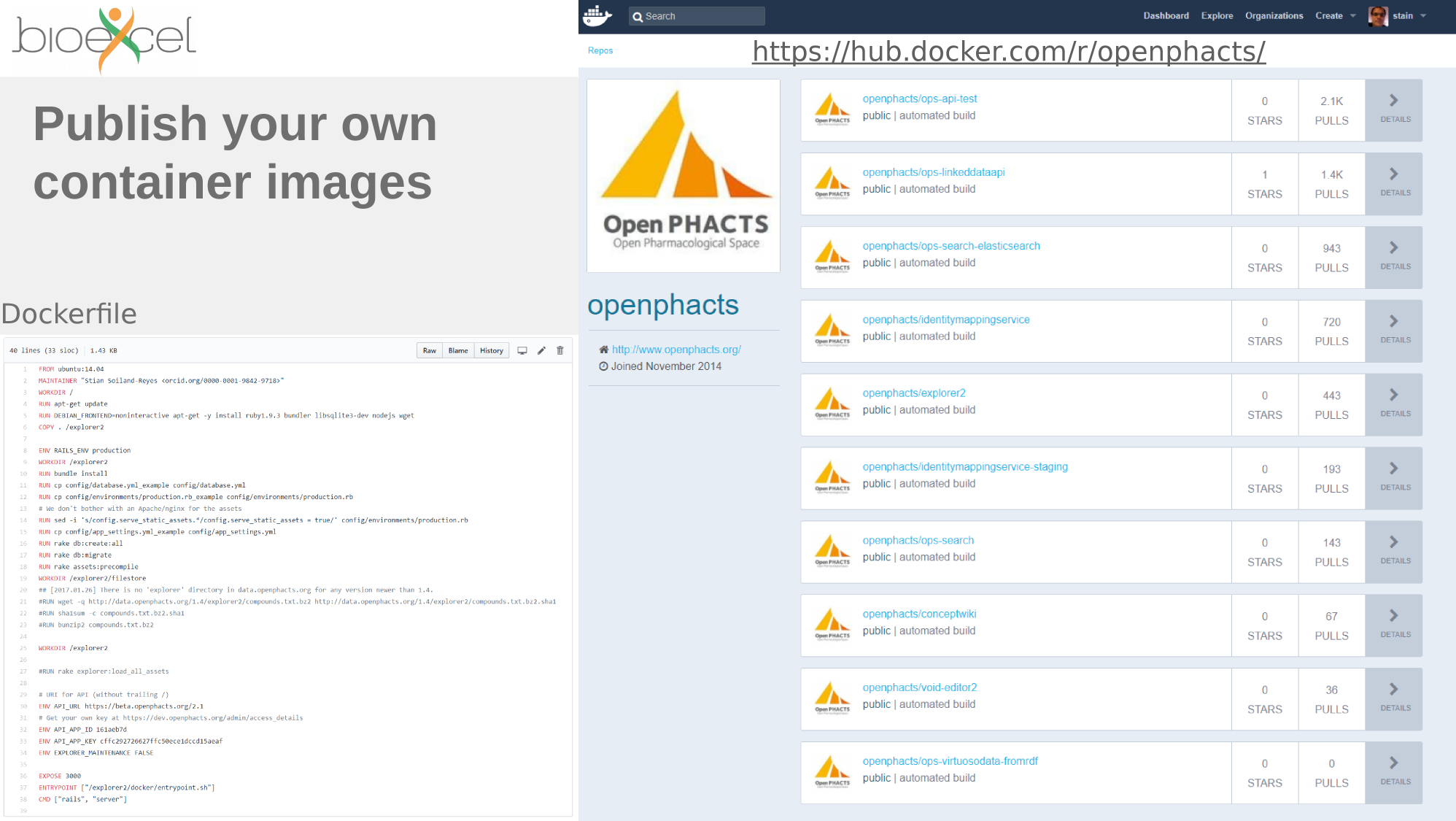




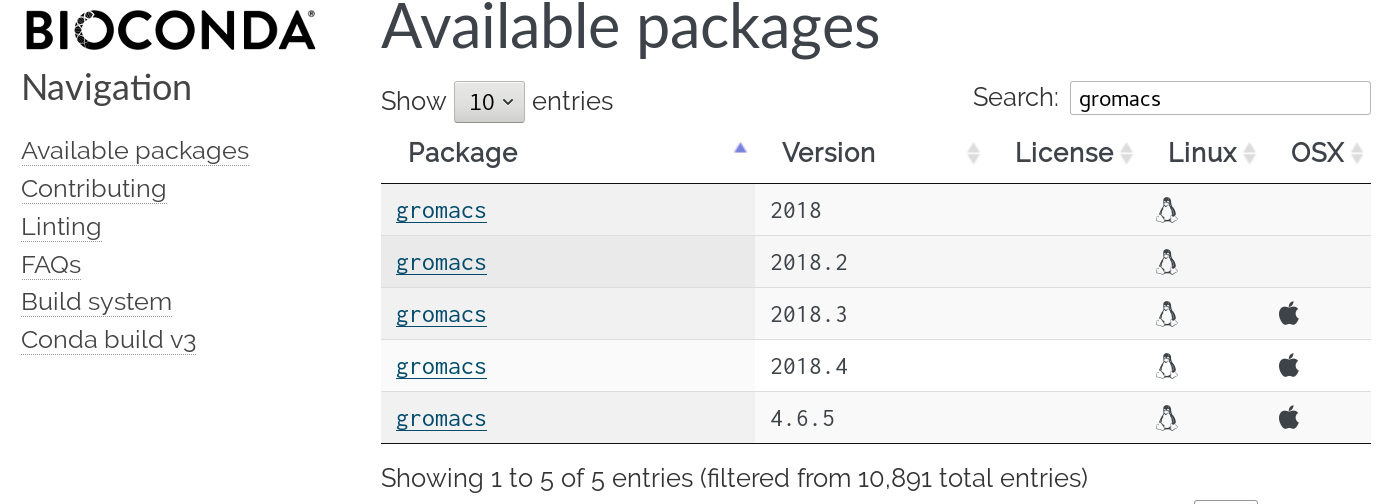
Repositories
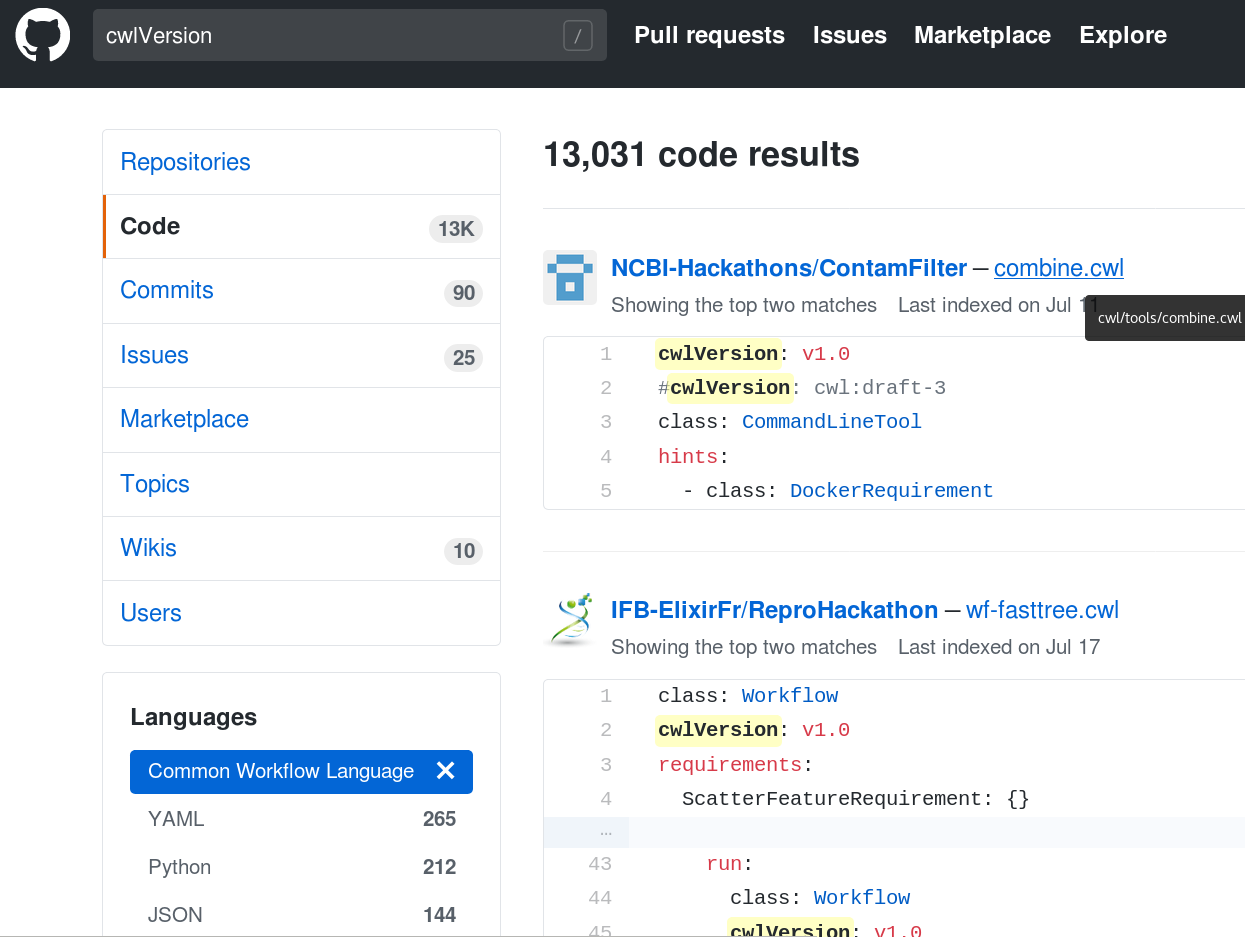
Sharing computational analyses
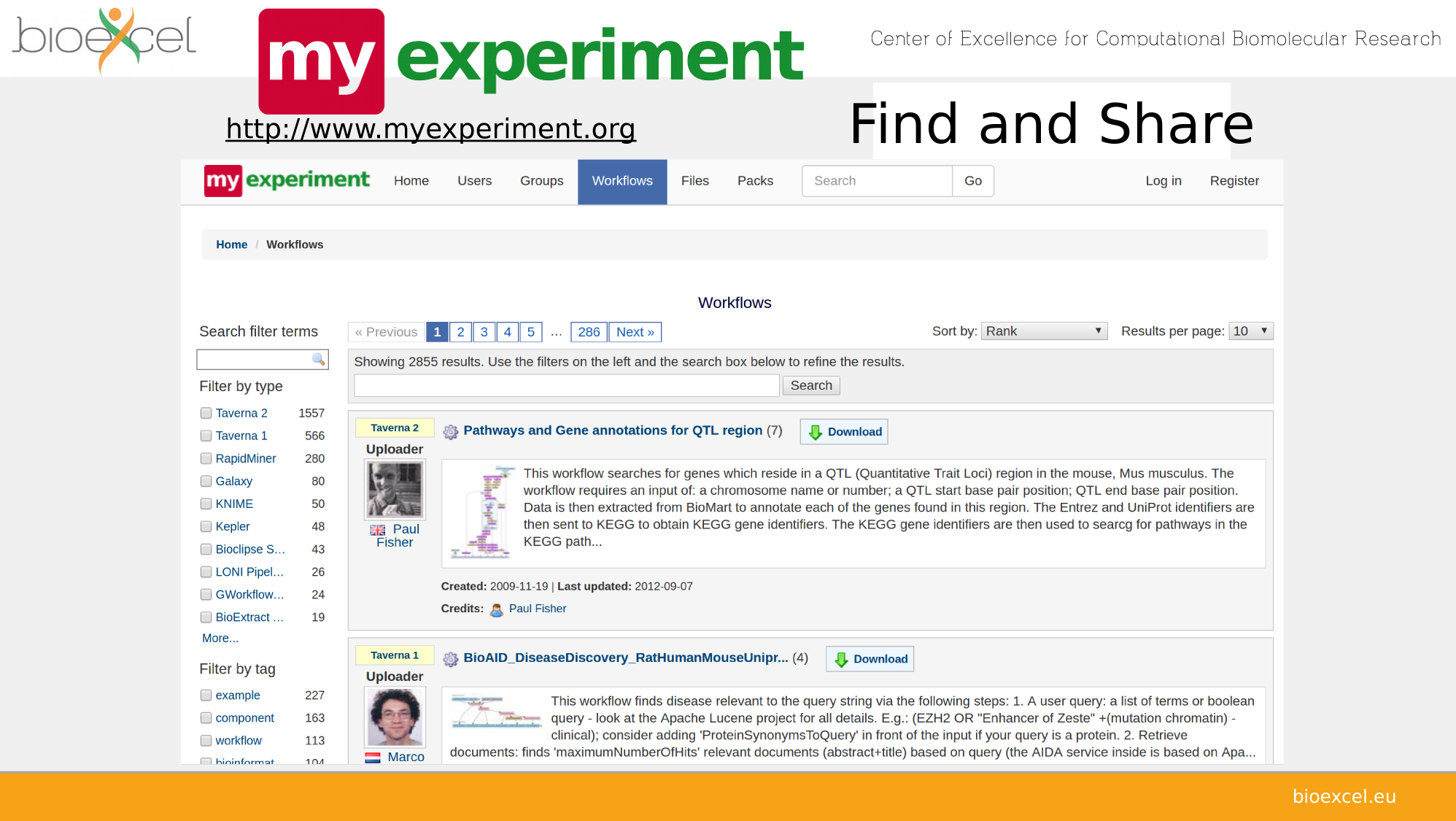
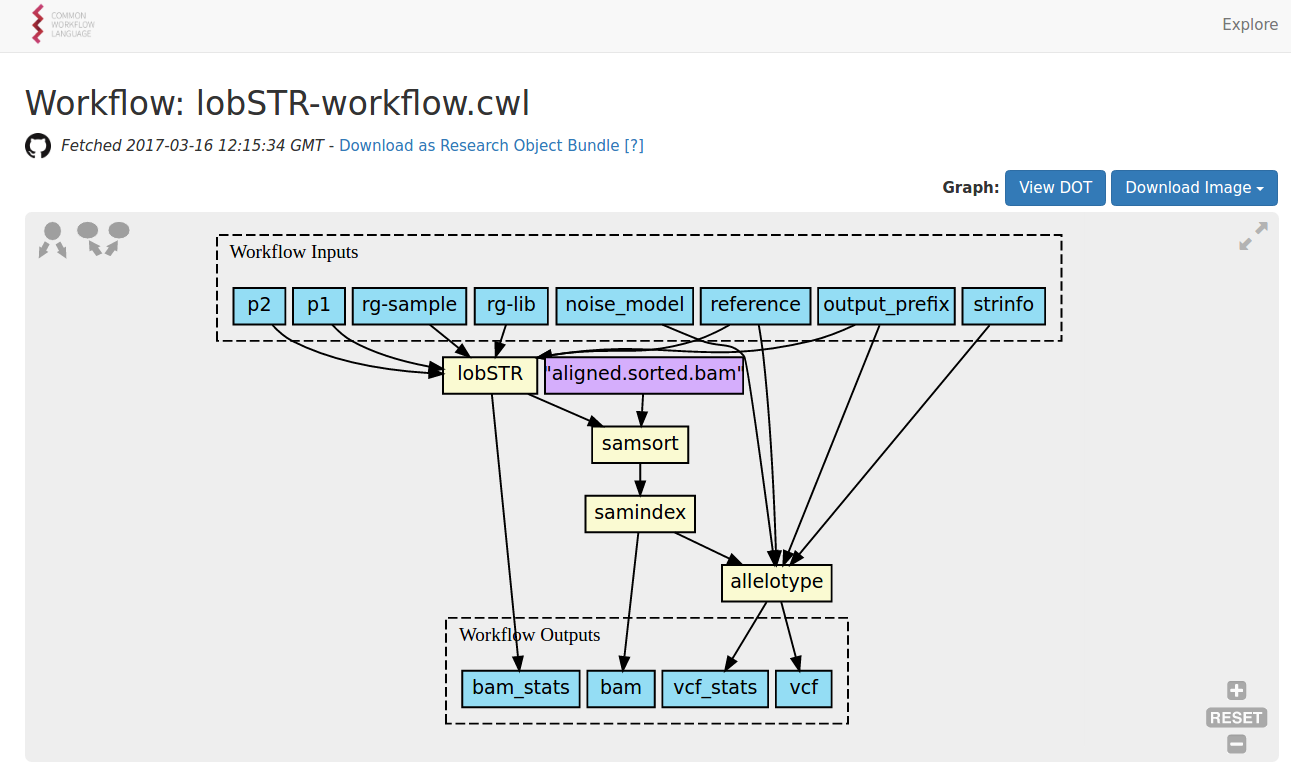
https://view.commonwl.org/workflows

Data repositories






Selecting a repository

Provenance
Who ran it?
When did it run?
Where did it run?
What workflow ran?
Which tool versions?
What data was created?
Copyright © 2013 W3C® (MIT, ERCIM, Keio, Beihang), All Rights Reserved. W3C liability, trademark and document use rules apply.
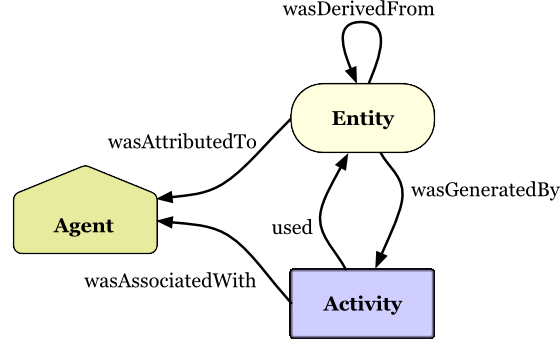
PROV Model Primer
W3C Working Group Note 30 April 2013

Khan et al,
Sharing interoperable workflow provenance: A review of best practices and their practical application in CWLProv
Submitted to GigaScience
https://doi.org/10.5281/zenodo.1966881


$ cwlprov --help
usage: cwlprov [-h] [--version] [--directory DIRECTORY] [--relative]
[--absolute] [--output OUTPUT] [--verbose] [--quiet] [--hints]
[--no-hints]
{validate,info,who,prov,inputs,outputs,run,runs,rerun,derived,runtimes}
...
cwlprov explores Research Objects containing provenance of Common Workflow
Language executions. <https://w3id.org/cwl/prov/>
commands:
{validate,info,who,prov,inputs,outputs,run,runs,rerun,derived,runtimes}
validate Validate the CWLProv Research Object
info show research object Metadata
who show Who ran the workflow
prov export workflow execution Provenance in PROV format
inputs list workflow/step Input files/values
outputs list workflow/step Output files/values
run show workflow Execution log
runs List all workflow executions in RO
rerun Rerun a workflow or step
derived list what was Derived from a data item, based on
activity usage/generation
runtimes calculate average step execution Runtimes


A Research Object bundles and relates digital resources of a scientific experiment or investigation:
Data used and results produced in experimental study
Methods employed to produce and analyse that data
Provenance and settings for the experiments
People involved in the investigation
Annotations about these resources, to improve understanding and interpretation
Research Object
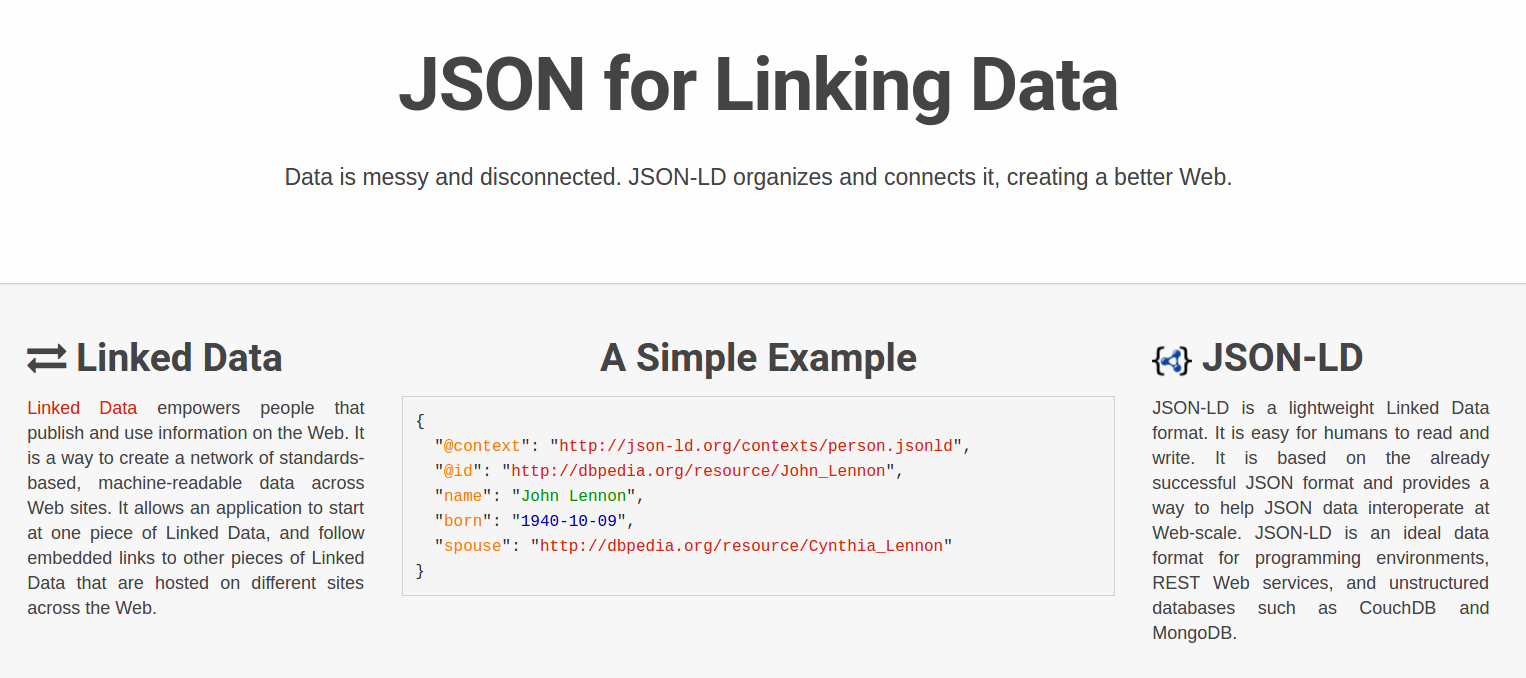
Who is using Research Objects?


















P2791


