RO-Crate, workflows and
FAIR Digital Objects
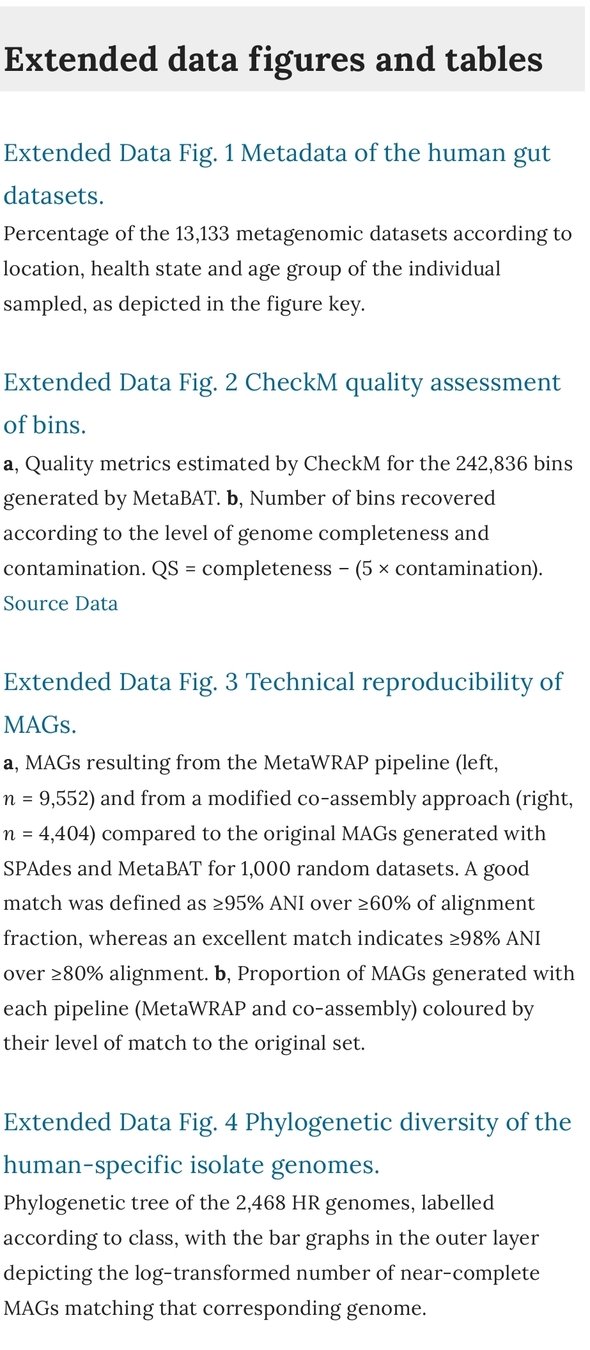
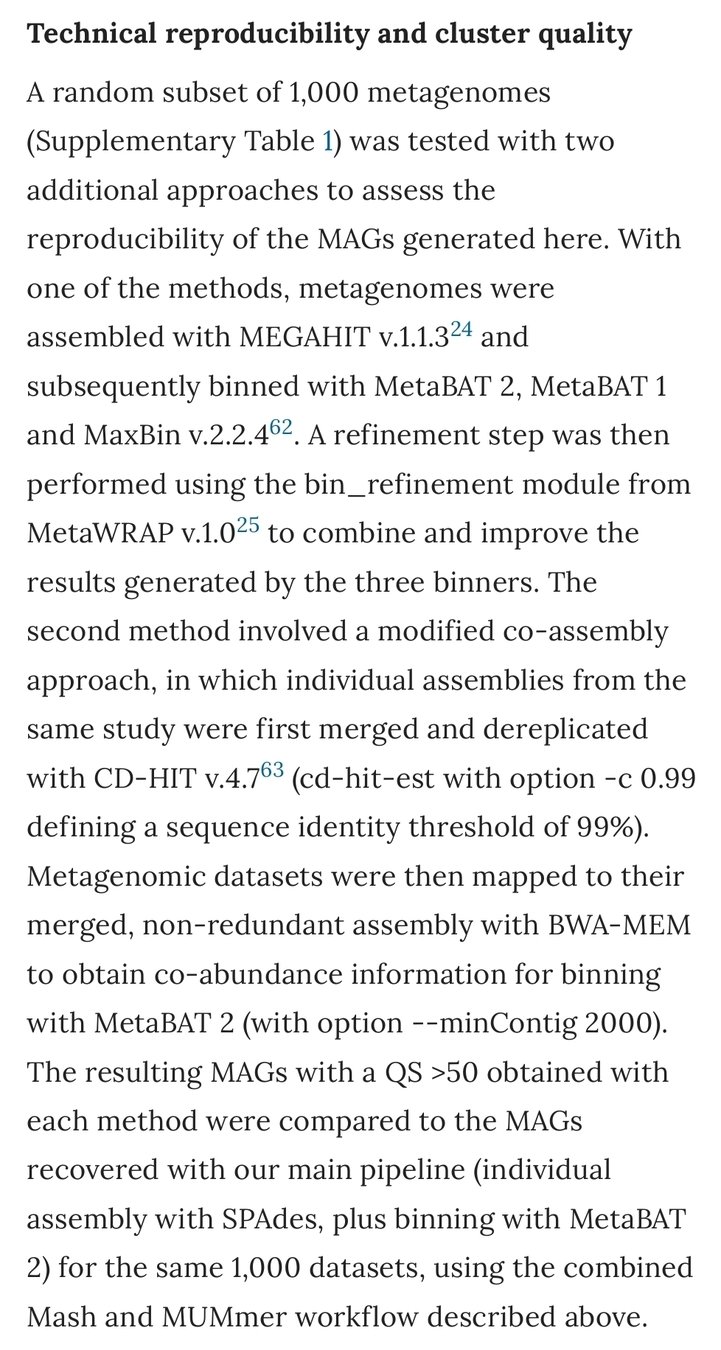
Stian Soiland-Reyes
eScience lab, The University of Manchester
INDElab, University of Amsterdam
FAIR Digital Object Forum
CWFR & FDO SEM meeting
2021-07-02
This work is licensed under a
Creative Commons Attribution 4.0 International License.










Findable
Accessible
Interoperable
Reusable

To be Findable:
F1. (meta)data are assigned a globally unique and persistent identifier
F2. data are described with rich metadata (defined by R1 below)
F3. metadata clearly and explicitly include the identifier of the data it describes
F4. (meta)data are registered or indexed in a searchable resource
To be Findable:
F1. (meta)data are assigned a globally unique and persistent identifier
F2. data are described with rich metadata (defined by R1 below)
F3. metadata clearly and explicitly include the identifier of the data it describes
F4. (meta)data are registered or indexed in a searchable resource
To be Accessible:
A1. (meta)data are retrievable by their identifier using a standardized communications protocol
A1.1 the protocol is open, free, and universally implementable
A1.2 the protocol allows for an authentication and authorization procedure, where necessary
A2. metadata are accessible, even when the data are no longer available
To be Findable:
F1. (meta)data are assigned a globally unique and persistent identifier
F2. data are described with rich metadata (defined by R1 below)
F3. metadata clearly and explicitly include the identifier of the data it describes
F4. (meta)data are registered or indexed in a searchable resource
To be Accessible:
A1. (meta)data are retrievable by their identifier using a standardized communications protocol
A1.1 the protocol is open, free, and universally implementable
A1.2 the protocol allows for an authentication and authorization procedure, where necessary
A2. metadata are accessible, even when the data are no longer available
To be Interoperable:
I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation.
I2. (meta)data use vocabularies that follow FAIR principles
I3. (meta)data include qualified references to other (meta)data
To be Findable:
F1. (meta)data are assigned a globally unique and persistent identifier
F2. data are described with rich metadata (defined by R1 below)
F3. metadata clearly and explicitly include the identifier of the data it describes
F4. (meta)data are registered or indexed in a searchable resource
To be Accessible:
A1. (meta)data are retrievable by their identifier using a standardized communications protocol
A1.1 the protocol is open, free, and universally implementable
A1.2 the protocol allows for an authentication and authorization procedure, where necessary
A2. metadata are accessible, even when the data are no longer available
To be Interoperable:
I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation.
I2. (meta)data use vocabularies that follow FAIR principles
I3. (meta)data include qualified references to other (meta)data
To be Reusable:
R1. meta(data) are richly described with a plurality of accurate and relevant attributes
R1.1. (meta)data are released with a clear and accessible data usage license
R1.2. (meta)data are associated with detailed provenance
R1.3. (meta)data meet domain-relevant community standards
To be Findable:
F1. (meta)data are assigned a globally unique and persistent identifier
F2. data are described with rich metadata (defined by R1 below)
F3. metadata clearly and explicitly include the identifier of the data it describes
F4. (meta)data are registered or indexed in a searchable resource
To be Accessible:
A1. (meta)data are retrievable by their identifier using a standardized communications protocol
A1.1 the protocol is open, free, and universally implementable
A1.2 the protocol allows for an authentication and authorization procedure, where necessary
A2. metadata are accessible, even when the data are no longer available
To be Interoperable:
I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation.
I2. (meta)data use vocabularies that follow FAIR principles
I3. (meta)data include qualified references to other (meta)data
To be Reusable:
R1. meta(data) are richly described with a plurality of accurate and relevant attributes
R1.1. (meta)data are released with a clear and accessible data usage license
R1.2. (meta)data are associated with detailed provenance
R1.3. (meta)data meet domain-relevant community standards


They ride with what I refer to as the four horsemen of the reproducibility apocalypse:
- Publication bias
- Low statistical power
- P-value hacking
- HARKing (hypothesizing after results are known)
Reproducibility?
Workflows
You can download our code from the URL supplied. Good luck downloading the only postdoc who can get it to run, though #overlyhonestmethods
— Ian Holmes (@ianholmes) January 8, 2013
Automation
– Automate computational aspects
– Repetitive pipelines, sweep campaigns
Scaling—compute cycles
– Make use of computational infrastructure
– Handle large data
Abstraction—people cycles
– Shield complexity and incompatibilities
– Report, re-usue, evolve, share, compare
– Repeat—Tweak—Repeat
– First-class commodities
Provenance—reporting
– Capture, report and utilize log and data lineage
– Auto-documentation
– Tracable evolution, audit, transparency
– Reproducible science
Findable
Accessible
Interoperable
Reusable
(Reproducible)
Why use workflows?
Adapted from Bertram Ludäscher (2015)
https://www.slideshare.net/ludaesch/works-2015provenancemileage
https://doi.org/10.1007/s13222-012-0100-z


cwlVersion: v1.0
class: Workflow
inputs:
inp: File
ex: string
outputs:
classout:
type: File
outputSource: compile/classfile
steps:
untar:
run: tar-param.cwl
in:
tarfile: inp
extractfile: ex
out: [example_out]
compile:
run: arguments.cwl
in:
src: untar/example_out
out: [classfile]

Nature 573, 149-150 (2019)
https://doi.org/10.1038/d41586-019-02619-z



cwlVersion: v1.0
class: Workflow
inputs:
toConvert: File
outputs:
converted:
type: File
outputSource: convertMethylation/converted
combined:
type: File
outputSource: mergeSymmetric/combined
steps:
convertMethylation:
run: interconverter.cwl
in:
toConvert: toConvert
out: [converted]
mergeSymmetric:
run: symmetriccpgs.cwl
in:
toCombine: convertMethylation/converted
out: [combined]
cwlVersion: v1.0
class: CommandLineTool
inputs:
toConvert:
type: File
inputBinding:
prefix: -i
outputs:
converted:
type: File
outputBinding:
glob: "*.meth"
baseCommand: interconverter.sh
arguments: ["-d", $(runtime.outdir)]
hints:
- class: DockerRequirement
dockerPull: "quay.io/neksa/screw-tool"
cwlVersion: v1.0
class: CommandLineTool
inputs:
toCombine:
type: File
inputBinding:
prefix: -i
outputs:
combined:
type: File
outputBinding:
glob: "*.sym"
baseCommand: symmetriccpgs.sh
arguments: ["-d", $(runtime.outdir)]
hints:
- class: DockerRequirement
dockerPull: "quay.io/neksa/screw-tool"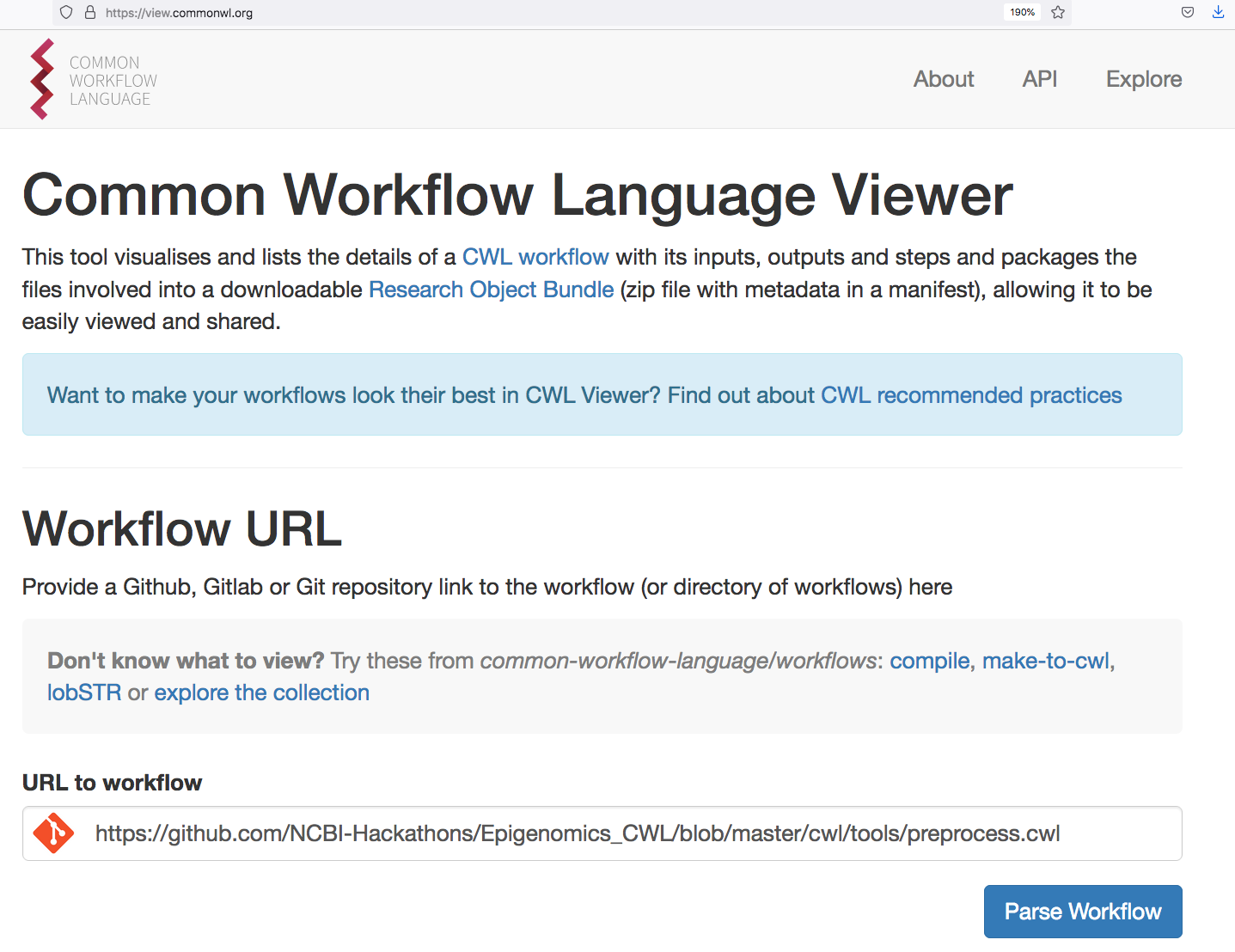
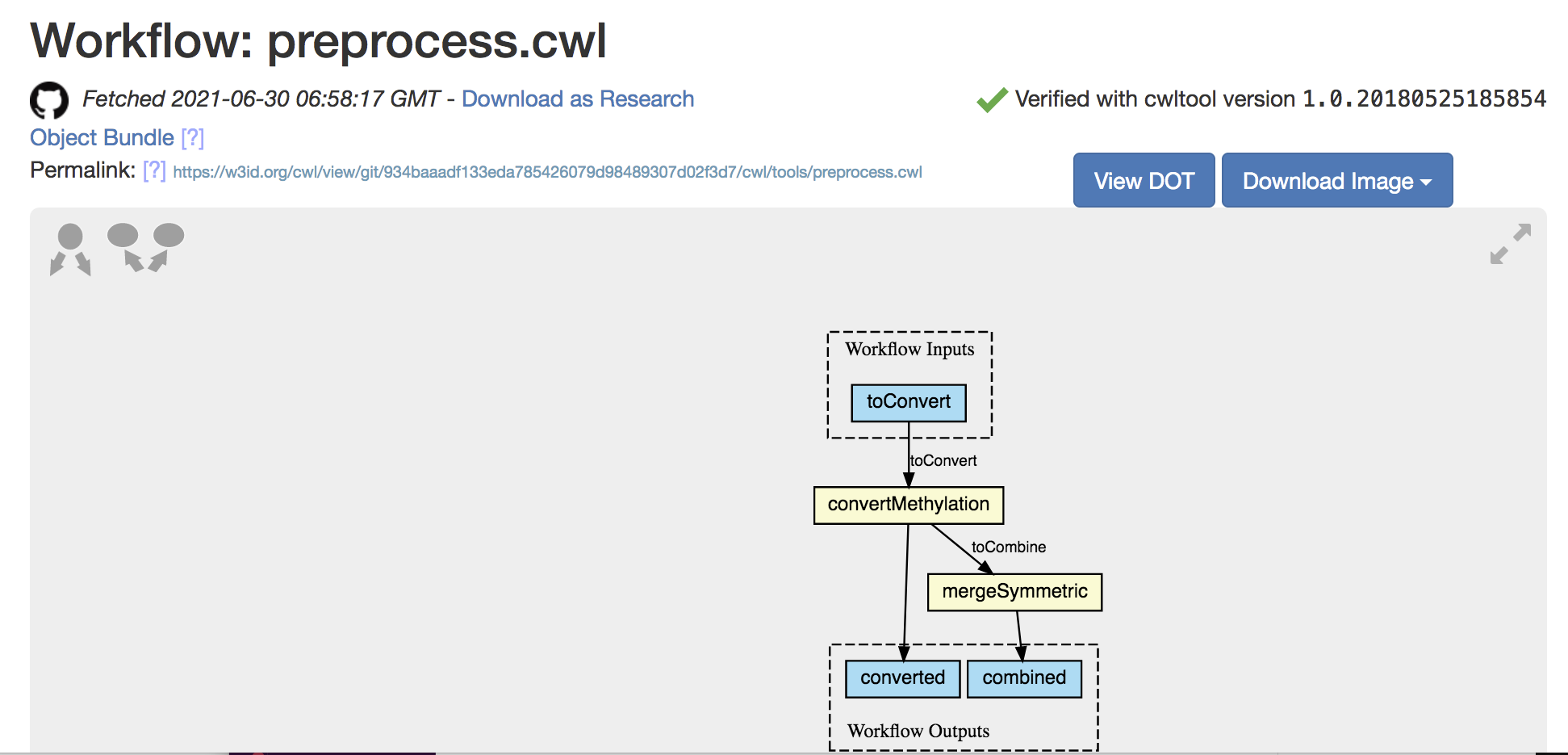

Digital Objects for CWL workflows



<https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/symmetriccpgs.cwl>
a cwl:CommandLineTool ;
cwl:arguments ( "-d" "$(runtime.outdir)" ) ;
cwl:baseCommand ( "symmetriccpgs.sh" ) ;
cwl:cwlVersion cwl:v1.0 ;
cwl:hints [ a cwl:DockerRequirement ;
DockerRequirement:dockerPull "quay.io/neksa/screw-tool"
] ;
cwl:inputs <https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/symmetriccpgs.cwl#toCombine> ;
cwl:outputs <https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/symmetriccpgs.cwl#combined> .
<https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/preprocess.cwl>
a cwl:Workflow ;
Workflow:steps <https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/preprocess.cwl#convertMethylation> , <https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/preprocess.cwl#mergeSymmetric> ;
cwl:cwlVersion cwl:v1.0 ;
cwl:inputs <https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/preprocess.cwl#toConvert> ;
cwl:outputs <https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/preprocess.cwl#combined> , <https://w3id.org/cwl/view/git/934baaadf133eda785426079d98489307d02f3d7/cwl/tools/preprocess.cwl#converted> .

cwlVersion: v1.0
class: Workflow
inputs:
toConvert: File
outputs:
converted:
type: File
outputSource: convertMethylation/converted
combined:
type: File
outputSource: mergeSymmetric/combined
steps:
convertMethylation:
run: interconverter.cwl
in:
toConvert: toConvert
out: [converted]
mergeSymmetric:
run: symmetriccpgs.cwl
in:
toCombine: convertMethylation/converted
out: [combined]



cwlVersion: v1.0
class: CommandLineTool
baseCommand: interconverter.sh
hints:
- class: DockerRequirement
dockerPull: "quay.io/neksa/screw-tool"
arguments: ["-d", $(runtime.outdir)]
inputs:
toConvert:
type: File
inputBinding:
prefix: -i
outputs:
converted:
type: File
outputBinding:
glob: "*.meth"cwlVersion: v1.0
class: CommandLineTool
baseCommand: symmetriccpgs.sh
arguments: ["-d", $(runtime.outdir)]
hints:
- class: DockerRequirement
dockerPull: "quay.io/neksa/screw-tool"
inputs:
toCombine:
type: File
inputBinding:
prefix: -i
outputs:
combined:
type: File
outputBinding:
glob: "*.sym"

?






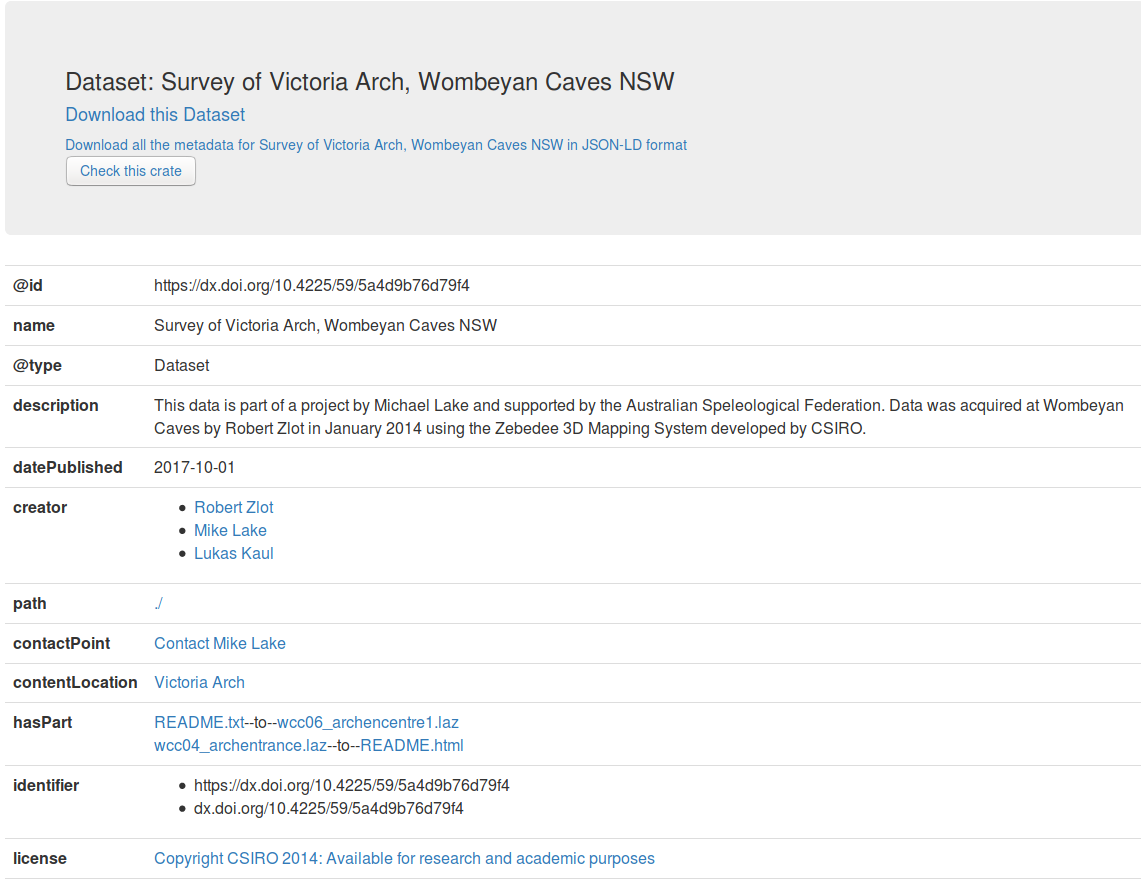
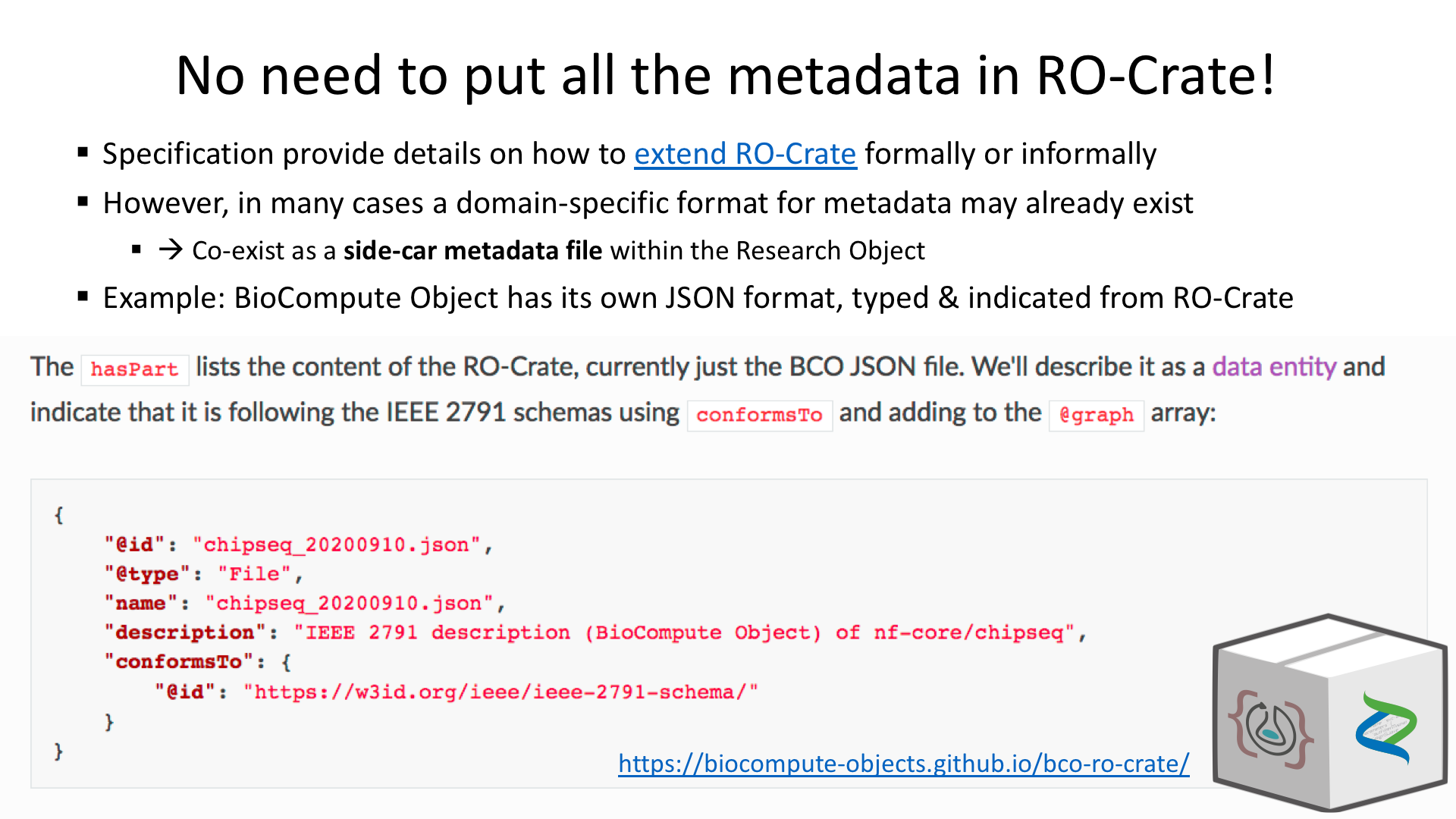
What is RO-Crate?

RO-Crate is method for self-decribed datasets as a digital object using a single Linked Data metadata document
Credit: Peter Sefton
Adapted from https://arkisto-platform.github.io/standards/ro-crate/
The dataset may contain any kind of
resource, about anything, in any format
as a file, URL or PID
Credit: Peter Sefton
Adapted from https://arkisto-platform.github.io/standards/ro-crate/

Each resource have a machine readable description in JSON-LD format
Credit: Peter Sefton
Adapted from https://arkisto-platform.github.io/standards/ro-crate/



A human-readable description/preview in an HTML file that lives alongside the metadata
Credit: Peter Sefton
Adapted from https://arkisto-platform.github.io/standards/ro-crate/
Provenance and workflow information can be included
– to assist in re-use of data and research processes
Credit: Peter Sefton
Adapted from https://arkisto-platform.github.io/standards/ro-crate/

RO-Crate Digital Objects may be packaged for distribution eg via Zip, Bagit and OCFL
– or simply be published on the Web
Credit: Peter Sefton
Adapted from https://arkisto-platform.github.io/standards/ro-crate/


Techie deep-dive!
Warning: JSON ahead


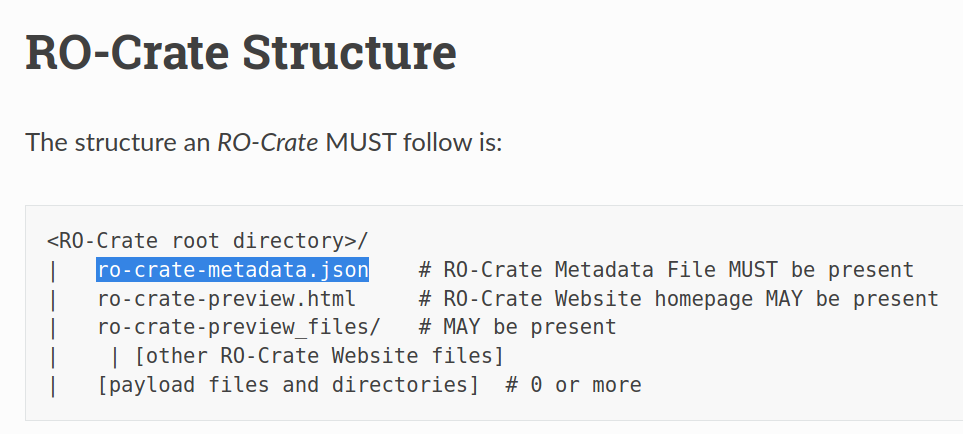

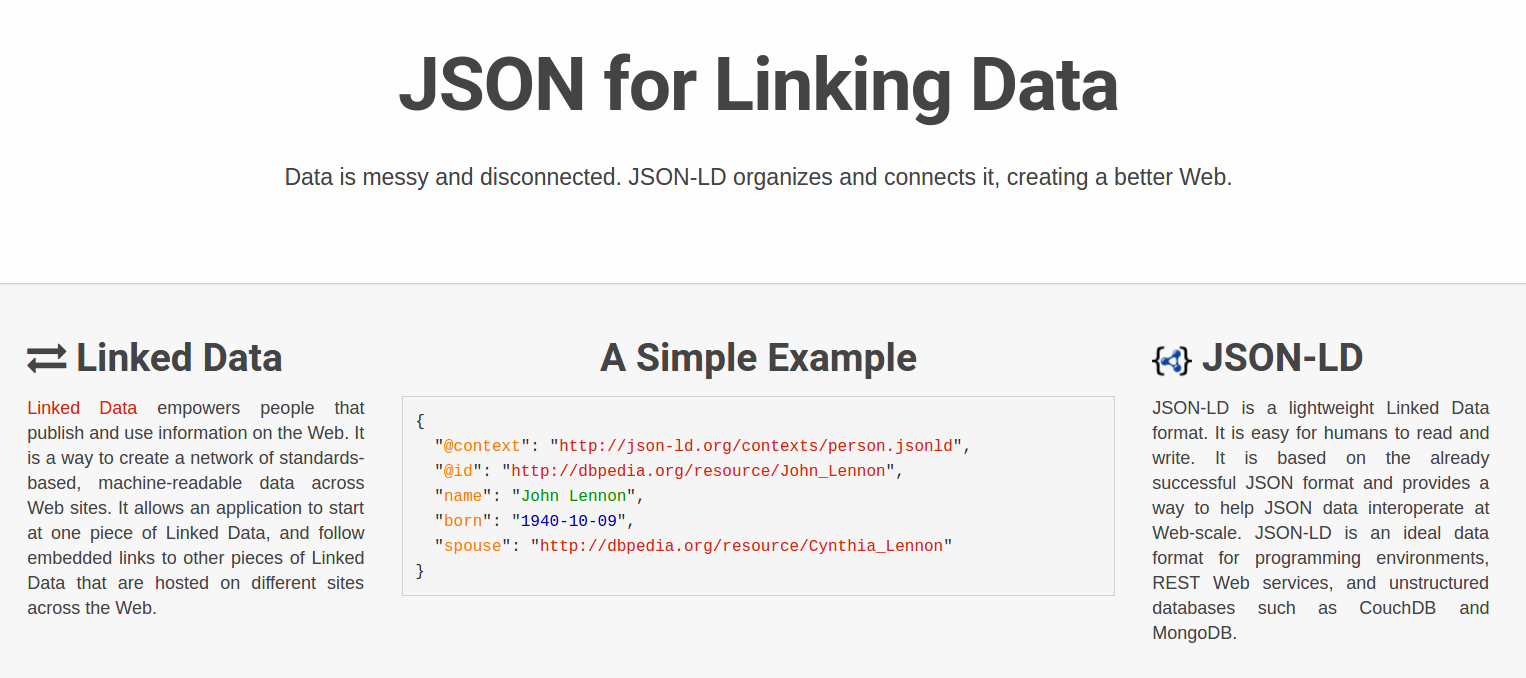
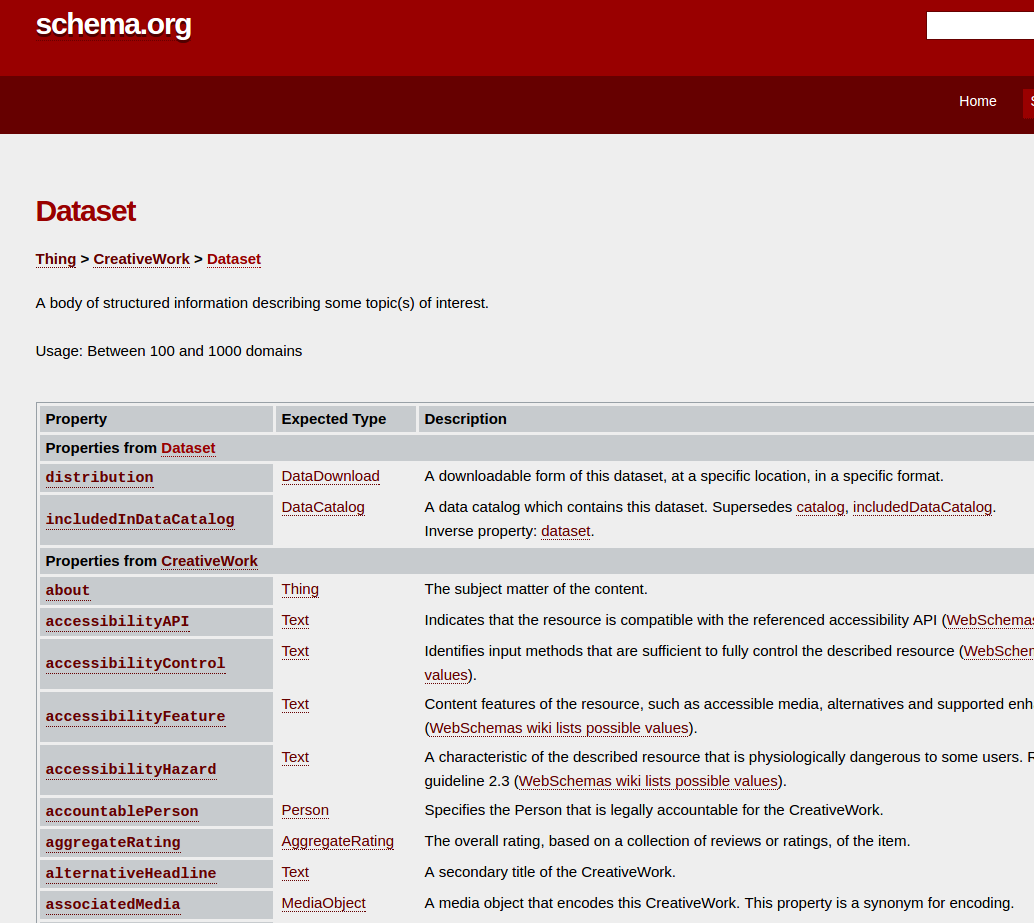


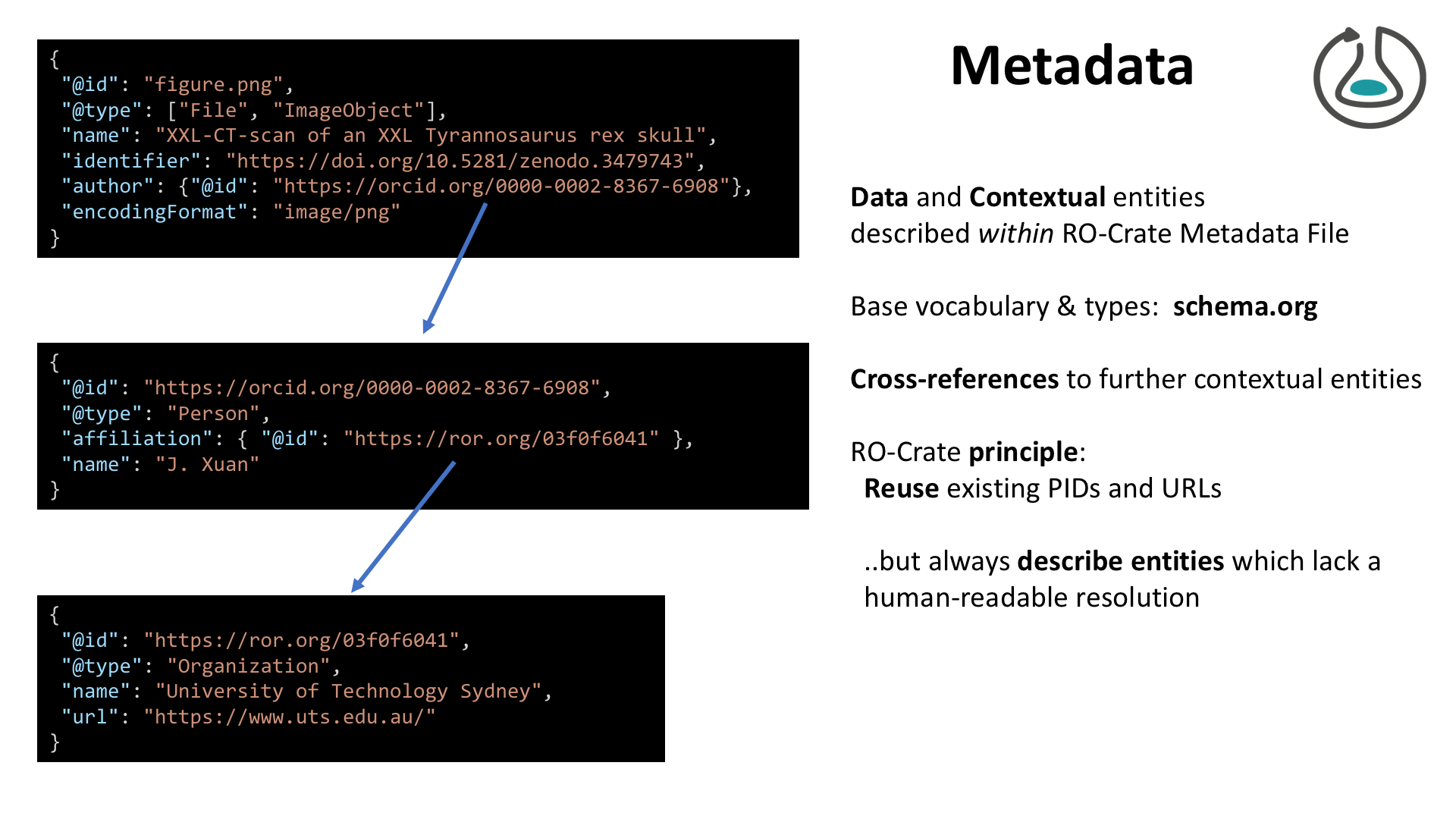

RO-Crate for data scientists
Credit: Marco La Rosa, Peter Sefton
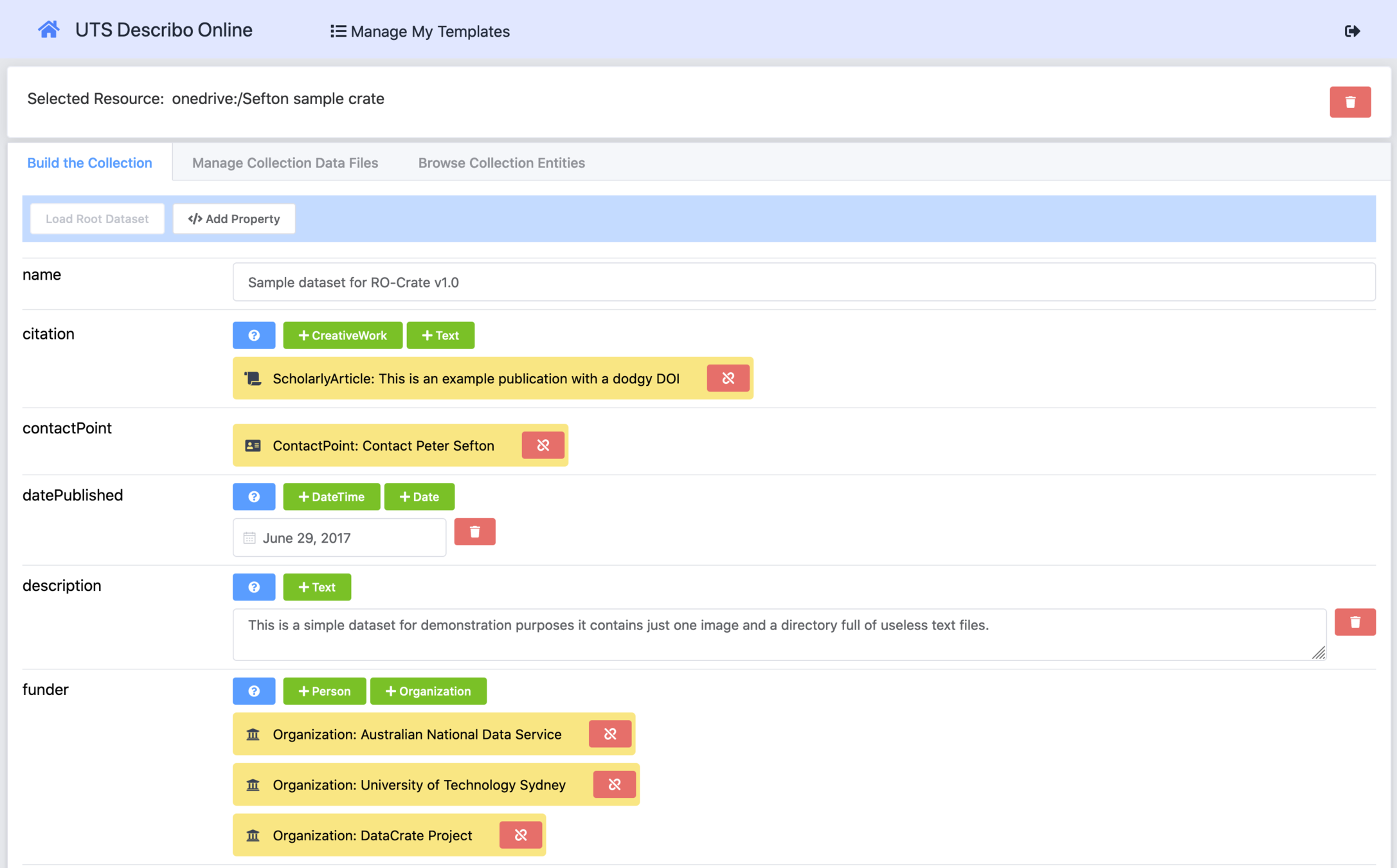
Making your own RO-Crate with Describo
FAIR is not just machine-readable!
RO-Crate for digital humanities
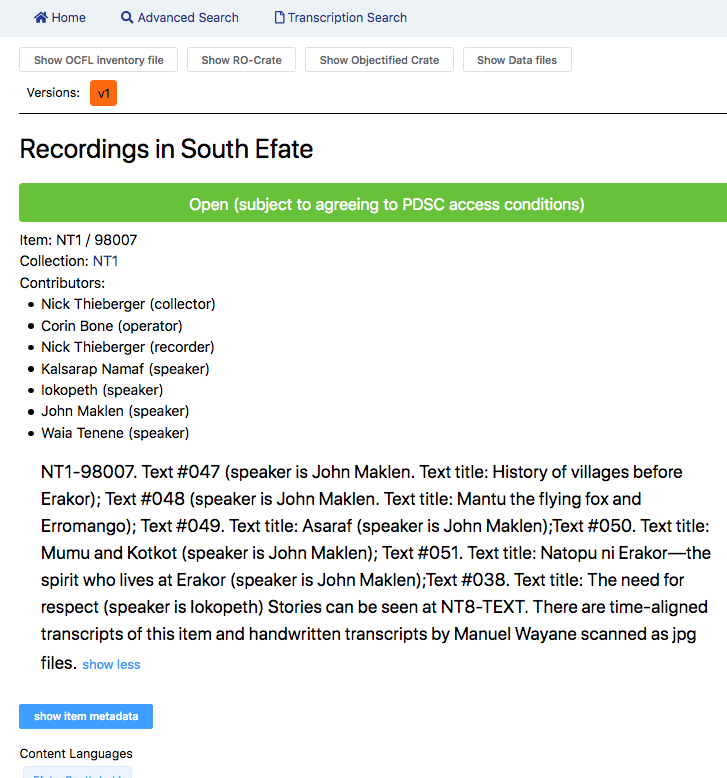
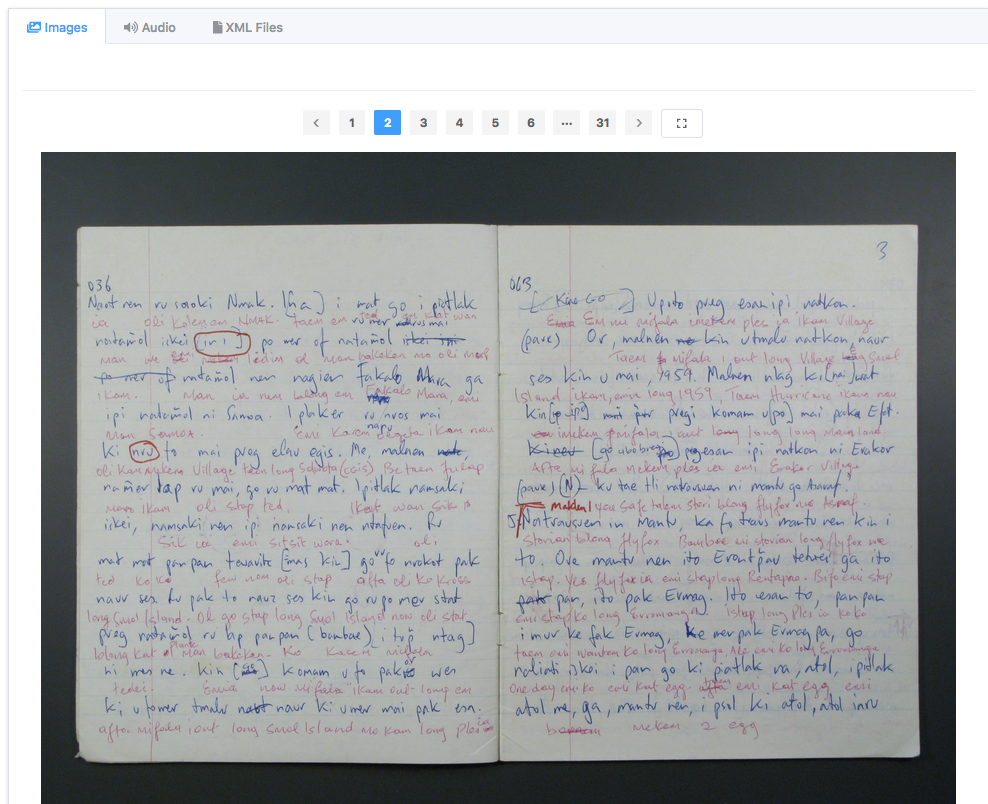

Capturing cultural heritage records
as RO-Crates
RO-Crate for repositories

RO-Crate as a
metadata archival format




Metadata held alongside hetereogeneous data
Exchange mechanism (import/export)
Avoid vendor lock-in



RO-Crate for
workflow descriptions

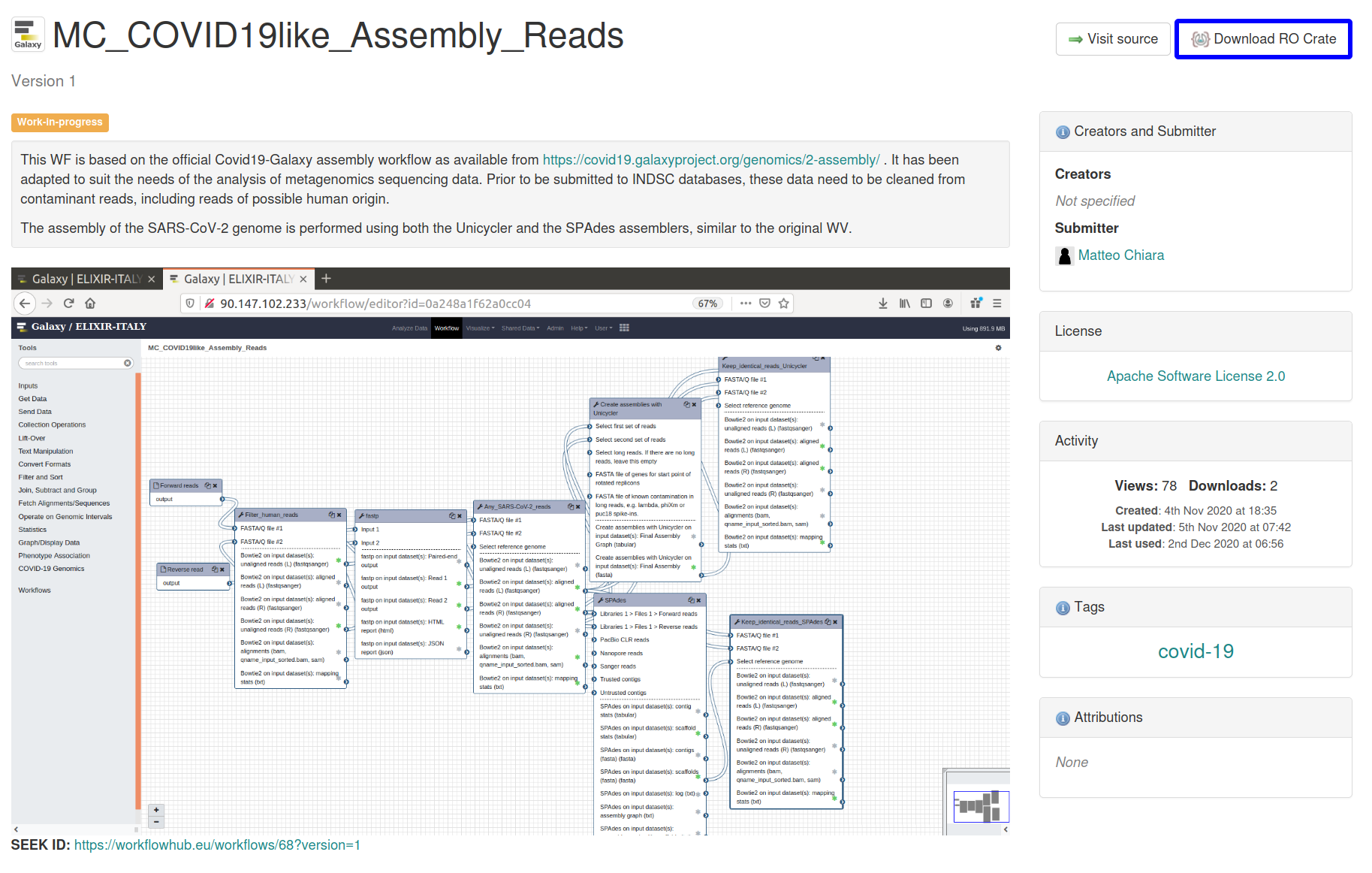


Describing workflows with RO-Crate






Describing workflows
with RO-Crate











Containers
Describe workflow
Tests
Registry
Workflows
Authors and contributors
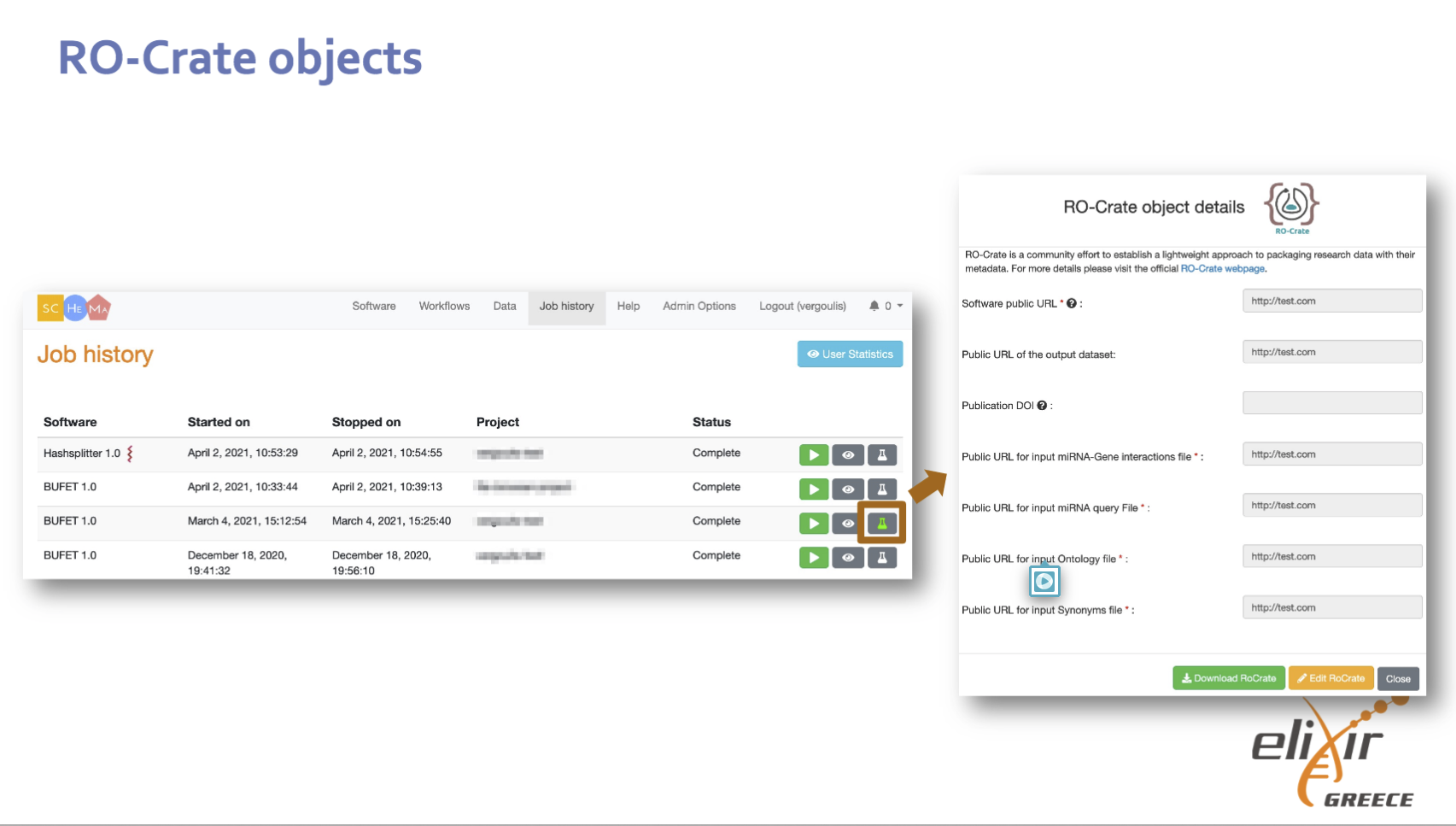
Credit: Thanasis Vergoulis
Executing Workflow RO-Crates

RO-Crate for
workflow test specifications
Credit: Simone Leo

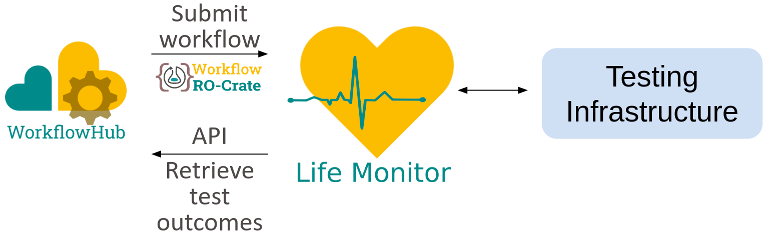
Workflow Testing RO-Crate
- Workflow definition (e.g. Galaxy, Snakemake)
- Test suite: Instances of Test definitions
- Binds to particular test engines, e.g. Planemo, Jenkins
RO-Crate for
computational tools




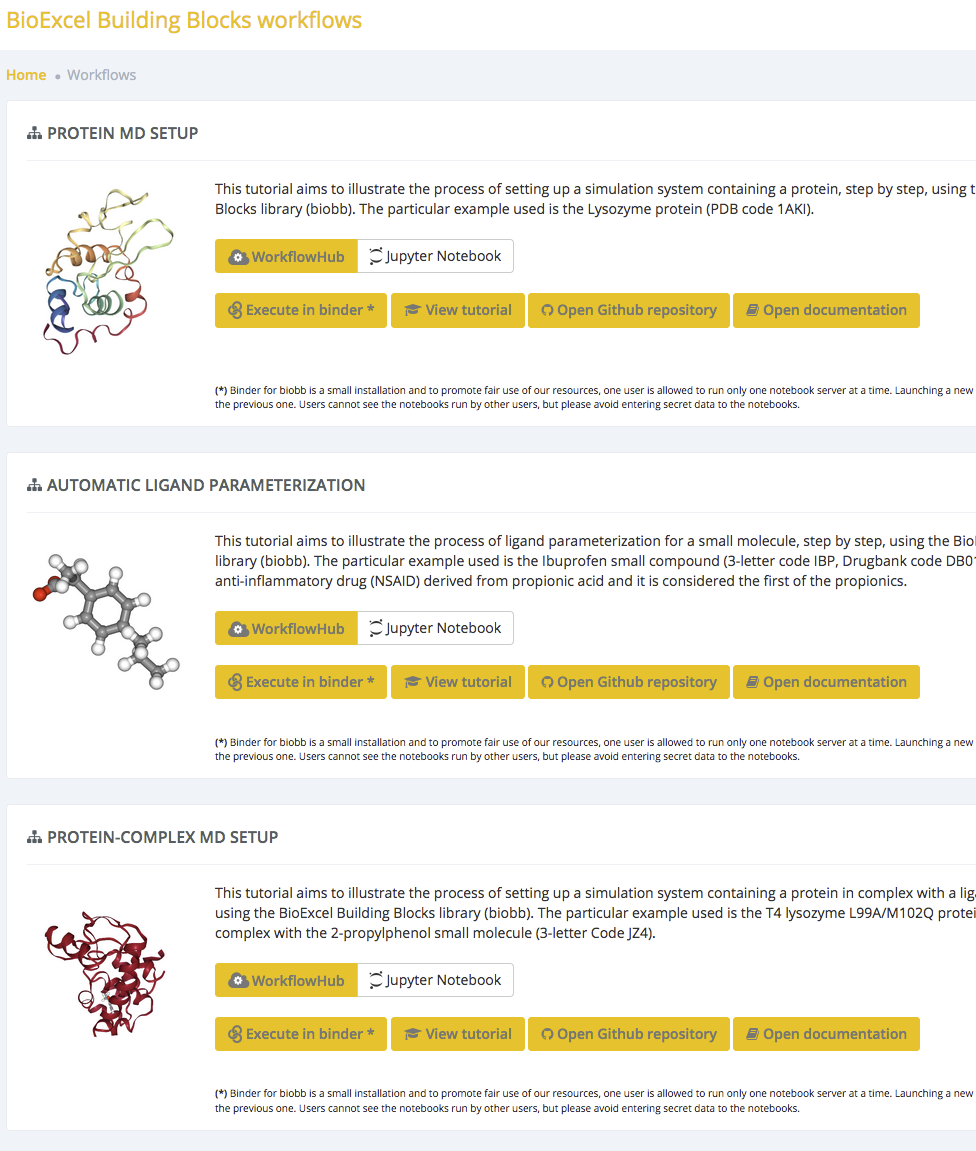




Making Canonical Workflow Building Blocks interoperable across workflow languages
RO-Crate for
workflow run provenance
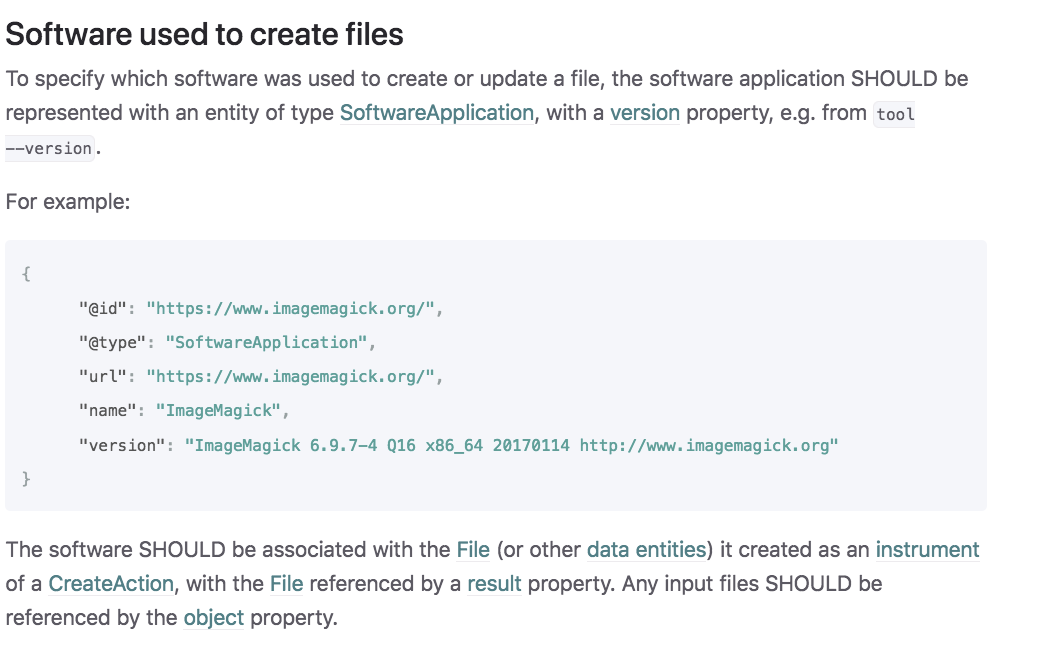
RO-Crate minimal provenance: Some software was used

Credit: José Mª Fernández, ELIXIR All Hands, 2021-06-11
What is needed for a Workflow Run RO-Crate?
-
Workflow language & version
-
Workflow engine & version (e.g. Toil)
-
Workflow definition
-
Input data (or pointers to such)
-
Parameters? What can be implicit and explicit? (see BCO?)
-
Tool Dependencies to install (mostly implied by CWL/Nextflow/Galaxy, but might need versions/repos)
-
Container platform requirement [e.g. Docker, Conda]
-
Operating system requirement
-
Hardware requirements (memory, CPU, GPU)
-
Equivalent of AWS cloud instance type sufficient?
-
-
Where to run/submit (e.g. usegalaxy.eu)
-
Explicit/resolved container IDs
-
Archive containers from Docker Hub (protect against image expiration)
-
...

Join discussion in the
Workflow Hub Club community!
https://about.workflowhub.eu/
--> Separation of concern
RO-Crate for
regulatory sciences
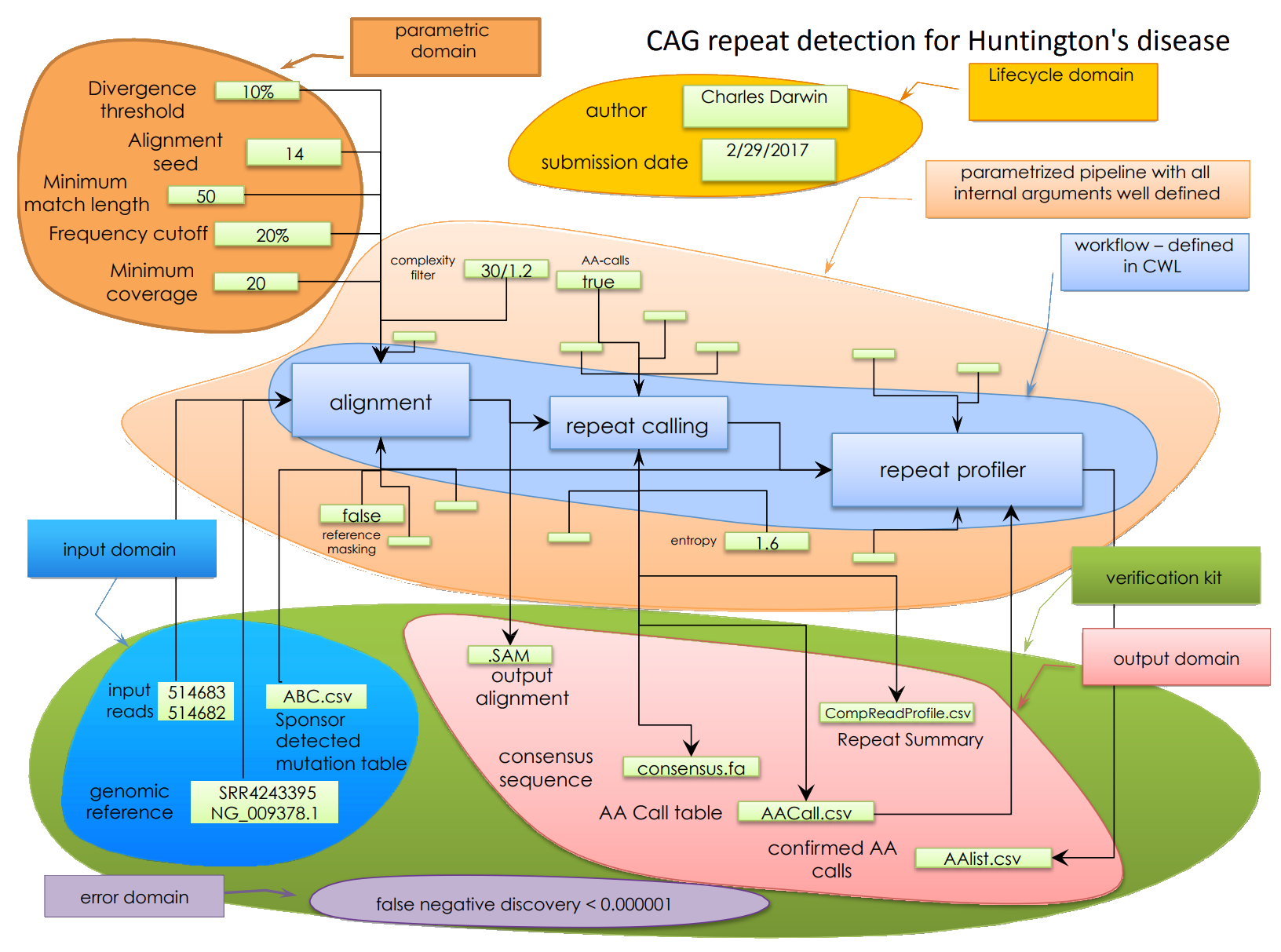




IEEE2791-2020

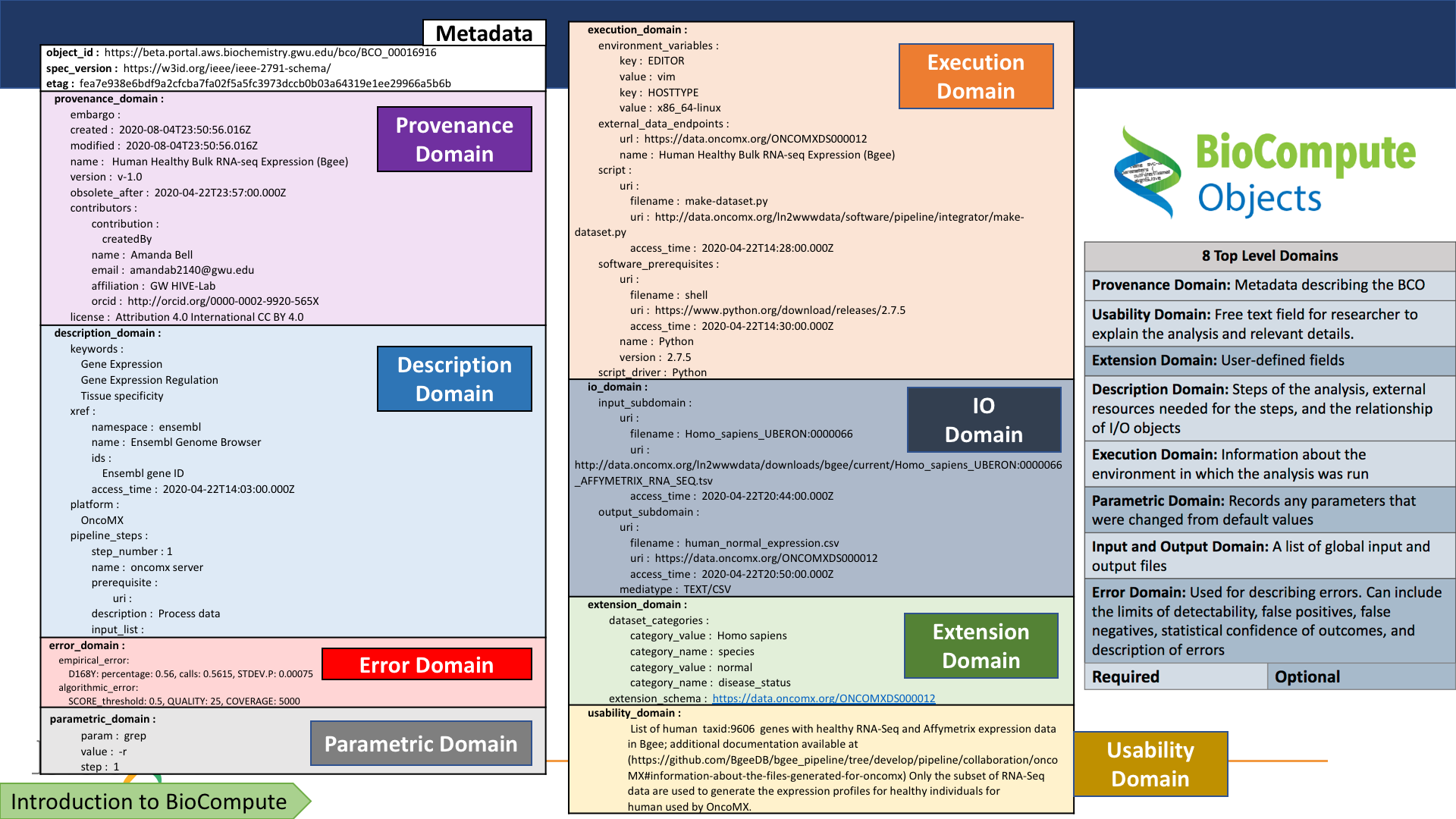


Alternate metadata views
Domain-specific explanation: BCO
General index: RO-Crate
ro-crate-metadata.json
{
"@context": [
"https://w3id.org/ro/crate/1.0/context",
{
"@vocab": "https://schema.org/"
}
],
"@graph": [
{
"@id": "ro-crate-metadata.json",
"@type": "CreativeWork",
"about": {
"@id": "./"
},
"identifier": "ro-crate-metadata.json",
"conformsTo": {
"@id": "https://w3id.org/ro/crate/1.0"
},
"license": {
"@id": "https://creativecommons.org/licenses/by-sa/3.0"
},
"description": "Made with Describo: https://uts-eresearch.github.io/describo/"
},
{
"@type": "Dataset",
"author": {
"@id": "https://orcid.org/0000-0001-9842-9718"
},
"citation": {
"@id": "https://doi.org/10.5281/zenodo.3966161"
},
"contactPoint": {
"@id": "https://github.com/biocompute-objects/bco-ro-example-chipseq/issues"
},
"datePublished": "2020-09-09T23:00:00.000Z",
"description": "Workflow run of a ChIP-seq peak-calling, QC and differential analysis pipeline",
"distribution": {
"@id": "https://github.com/biocompute-objects/bco-ro-example-chipseq/archive/main.zip"
},
"hasPart": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
},
{
"@id": "chipseq_20200910.json"
},
{
"@id": "results/"
},
{
"@id": "nextflow.log"
},
{
"@id": ".nextflow.log"
}
],
"license": {
"@id": "https://spdx.org/licenses/CC0-1.0"
},
"name": "Workflow run of nf-core/chipseq",
"publisher": {
"@id": "https://biocomputeobject.org/"
},
"@id": "./"
},
{
"@type": "File",
"dateModified": "2020-09-10T13:10:50.246Z",
"name": ".nextflow.log",
"@reverse": {
"hasPart": [
{
"@id": "./"
}
]
},
"@id": ".nextflow.log"
},
{
"@type": "File",
"conformsTo": {
"@id": "https://w3id.org/ieee/ieee-2791-schema/"
},
"dateModified": "2020-09-10T13:50:02.378Z",
"identifier": {
"@id": "urn:uuid:dc308d7c-7949-446a-9c39-511b8ab40caf"
},
"license": {
"@id": "https://spdx.org/licenses/CC0-1.0"
},
"name": "chipseq_20200910.json",
"description": "IEEE 2791 description",
"@reverse": {
"hasPart": [
{
"@id": "./"
}
]
},
"@id": "chipseq_20200910.json"
},
{
"@type": "Organization",
"description": " Two non-overlapping entities work in parallel to help drive BioCompute, the IEEE 2791-2020 Standard, and a Public Private Partnership. Leadership for the Public Private Partnership consists of an Executive Steering Committee and a Technical Steering Committee. The schema that is referenced by the current draft of the IEEE standard is maintained by an IEEE GitLab repository. ",
"name": "BioCompute Objects",
"@reverse": {
"publisher": [
{
"@id": "./"
}
]
},
"@id": "https://biocomputeobject.org/"
},
{
"@type": "ScholarlyArticle",
"name": "nf-core/chipseq: nf-core/chipseq v1.2.1 - Platinum Mole",
"@reverse": {
"citation": [
{
"@id": "./"
},
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "https://doi.org/10.5281/zenodo.3966161"
},
{
"@type": "CreativeWork",
"identifier": "https://spdx.org/licenses/MIT",
"name": "MIT License",
"@reverse": {
"license": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "https://github.com/nf-core/chipseq/blob/1.2.1/LICENSE"
},
{
"@type": "CreativeWork",
"description": "\nMIT License\n\nCopyright (c) 2018 nf-core\n\nPermission is hereby granted, free of charge, to any person obtaining a copy\nof this software and associated documentation files (the \"Software\"), to deal\nin the Software without restriction, including without limitation the rights\nto use, copy, modify, merge, publish, distribute, sublicense, and/or sell\ncopies of the Software, and to permit persons to whom the Software is\nfurnished to do so, subject to the following conditions:\n\nThe above copyright notice and this permission notice shall be included in all\ncopies or substantial portions of the Software.\n\nTHE SOFTWARE IS PROVIDED \"AS IS\", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR\nIMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,\nFITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE\nAUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER\nLIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,\nOUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE\nSOFTWARE.",
"name": "MIT License",
"@reverse": {
"license": [
{
"@id": "results/"
}
]
},
"@id": "https://github.com/nf-core/test-datasets/blob/atacseq/LICENSE"
},
{
"@type": "DataDownload",
"path": "https://github.com/biocompute-objects/bco-ro-example-chipseq/archive/main.zip",
"license": {
"@id": "https://spdx.org/licenses/CC0-1.0"
},
"name": "GitHub download of biocompute-objects/bco-ro-example-chipseq",
"@reverse": {
"distribution": [
{
"@id": "./"
}
]
},
"@id": "https://github.com/biocompute-objects/bco-ro-example-chipseq/archive/main.zip"
},
{
"@type": "ContactPoint",
"name": " bco-ro-example-chipseq GitHub issue tracker",
"url": "https://github.com/biocompute-objects/bco-ro-example-chipseq/issues",
"@reverse": {
"contactPoint": [
{
"@id": "./"
}
]
},
"@id": "https://github.com/biocompute-objects/bco-ro-example-chipseq/issues"
},
{
"@type": "Person",
"name": "Stian Soiland-Reyes",
"@reverse": {
"author": [
{
"@id": "./"
},
{
"@id": "results/"
}
]
},
"@id": "https://orcid.org/0000-0001-9842-9718"
},
{
"@type": [
"ComputationalWorkflow",
"File"
],
"author": [
{
"@id": "#714de175-aa77-47f1-9f99-6a4fba65530a"
},
{
"@id": "#bfb876e7-e767-4209-ad66-e1e1379c249f"
},
{
"@id": "#0164006f-bd58-4ebc-9a50-b8bd4ac3025c"
},
{
"@id": "#556c747c-376a-4a85-82a1-9b99520d24fd"
},
{
"@id": "#93c23523-03b5-41dc-be4c-6a9a2e0e221d"
},
{
"@id": "#781b9b5a-dc06-4709-8f14-65ee08b8c543"
},
{
"@id": "#f652b13e-0ba2-4394-a990-7304f54c7b9a"
},
{
"@id": "#a58abf42-751d-49bd-a477-1d5065ac70c6"
},
{
"@id": "#e11af59b-8e24-4cc8-8f5e-cef411ab0823"
},
{
"@id": "#f262954b-a218-480d-8a01-0e0b1ca20ffc"
},
{
"@id": "#64bd387d-60ad-4df8-804e-1f6b9ea72de5"
}
],
"citation": {
"@id": "https://doi.org/10.5281/zenodo.3966161"
},
"description": "nfcore/chipseq is a bioinformatics analysis pipeline used for Chromatin ImmunopreciPitation sequencing (ChIP-seq) data",
"license": {
"@id": "https://github.com/nf-core/chipseq/blob/1.2.1/LICENSE"
},
"name": "nf-core/chipseq",
"@reverse": {
"hasPart": [
{
"@id": "./"
}
],
"about": [
{
"@id": "results/pipeline_info/pipeline_dag.svg"
},
{
"@id": "#fcb32545-04bd-474d-9b6e-0fb7321c38b4"
}
]
},
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
},
{
"@type": "File",
"creator": {
"@id": "#db65dfb7-4867-400e-a12f-a1652d46a333"
},
"dateModified": "2020-09-10T13:10:50.250Z",
"name": "nextflow.log",
"@reverse": {
"hasPart": [
{
"@id": "./"
}
]
},
"@id": "nextflow.log"
},
{
"@type": "Dataset",
"author": {
"@id": "https://orcid.org/0000-0001-9842-9718"
},
"creator": {
"@id": "#db65dfb7-4867-400e-a12f-a1652d46a333"
},
"dateModified": "2020-09-10T13:20:49.143Z",
"description": "Nextflow outputs from examplar run of nf-core/ pipeline workflow.",
"hasPart": [
{
"@id": "results/bwa/"
},
{
"@id": "results/fastqc/"
},
{
"@id": "results/genome/"
},
{
"@id": "results/igv/"
},
{
"@id": "results/multiqc/"
},
{
"@id": "results/pipeline_info/"
},
{
"@id": "results/trim_galore/"
}
],
"license": {
"@id": "https://github.com/nf-core/test-datasets/blob/atacseq/LICENSE"
},
"name": "results",
"@reverse": {
"hasPart": [
{
"@id": "./"
}
]
},
"@id": "results/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:00:09.238Z",
"hasPart": {
"@id": "results/bwa/mergedLibrary/"
},
"name": "bwa",
"@reverse": {
"hasPart": [
{
"@id": "results/"
}
]
},
"@id": "results/bwa/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:02:59.495Z",
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/bigwig/"
},
{
"@id": "results/bwa/mergedLibrary/deepTools/"
},
{
"@id": "results/bwa/mergedLibrary/macs/"
},
{
"@id": "results/bwa/mergedLibrary/phantompeakqualtools/"
},
{
"@id": "results/bwa/mergedLibrary/picard_metrics/"
}
],
"name": "mergedLibrary",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/"
}
]
},
"@id": "results/bwa/mergedLibrary/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:04:31.692Z",
"hasPart": {
"@id": "results/bwa/mergedLibrary/bigwig/scale/"
},
"name": "bigwig",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/"
}
]
},
"@id": "results/bwa/mergedLibrary/bigwig/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:04:31.696Z",
"name": "scale",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/bigwig/"
}
]
},
"@id": "results/bwa/mergedLibrary/bigwig/scale/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:11:43.943Z",
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/deepTools/plotFingerprint/"
},
{
"@id": "results/bwa/mergedLibrary/deepTools/plotProfile/"
}
],
"name": "deepTools",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/"
}
]
},
"@id": "results/bwa/mergedLibrary/deepTools/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:05:17.700Z",
"name": "plotFingerprint",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/deepTools/"
}
]
},
"@id": "results/bwa/mergedLibrary/deepTools/plotFingerprint/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:26:12.375Z",
"name": "plotProfile",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/deepTools/"
}
]
},
"@id": "results/bwa/mergedLibrary/deepTools/plotProfile/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:02:33.471Z",
"name": "macs",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/"
}
]
},
"@id": "results/bwa/mergedLibrary/macs/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:04:26.336Z",
"name": "phantompeakqualtools",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/"
}
]
},
"@id": "results/bwa/mergedLibrary/phantompeakqualtools/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:04:45.952Z",
"name": "picard_metrics",
"@reverse": {
"hasPart": [
{
"@id": "results/bwa/mergedLibrary/"
}
]
},
"@id": "results/bwa/mergedLibrary/picard_metrics/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T11:58:56.905Z",
"hasPart": {
"@id": "results/fastqc/zips/"
},
"name": "fastqc",
"@reverse": {
"hasPart": [
{
"@id": "results/"
}
]
},
"@id": "results/fastqc/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T11:58:56.909Z",
"name": "zips",
"@reverse": {
"hasPart": [
{
"@id": "results/fastqc/"
}
]
},
"@id": "results/fastqc/zips/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T11:56:45.292Z",
"hasPart": {
"@id": "results/genome/genome.fa"
},
"name": "genome",
"@reverse": {
"hasPart": [
{
"@id": "results/"
}
]
},
"@id": "results/genome/"
},
{
"@type": "File",
"dateModified": "2020-09-10T11:56:45.324Z",
"name": "genome.fa",
"@reverse": {
"hasPart": [
{
"@id": "results/genome/"
}
]
},
"@id": "results/genome/genome.fa"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:26:50.263Z",
"hasPart": {
"@id": "results/igv/broadPeak/"
},
"name": "igv",
"@reverse": {
"hasPart": [
{
"@id": "results/"
}
]
},
"@id": "results/igv/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:26:50.267Z",
"hasPart": {
"@id": "results/igv/broadPeak/igv_session.xml"
},
"name": "broadPeak",
"@reverse": {
"hasPart": [
{
"@id": "results/igv/"
}
]
},
"@id": "results/igv/broadPeak/"
},
{
"@type": "File",
"dateModified": "2020-09-10T12:26:50.267Z",
"name": "igv_session.xml",
"@reverse": {
"hasPart": [
{
"@id": "results/igv/broadPeak/"
}
]
},
"@id": "results/igv/broadPeak/igv_session.xml"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:26:59.183Z",
"hasPart": {
"@id": "results/multiqc/broadPeak/"
},
"name": "multiqc",
"@reverse": {
"hasPart": [
{
"@id": "results/"
}
]
},
"@id": "results/multiqc/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:26:59.183Z",
"hasPart": [
{
"@id": "results/multiqc/broadPeak/multiqc_data/"
},
{
"@id": "results/multiqc/broadPeak/multiqc_report.html"
}
],
"name": "broadPeak",
"@reverse": {
"hasPart": [
{
"@id": "results/multiqc/"
}
]
},
"@id": "results/multiqc/broadPeak/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T12:26:59.207Z",
"name": "multiqc_data",
"@reverse": {
"hasPart": [
{
"@id": "results/multiqc/broadPeak/"
}
]
},
"@id": "results/multiqc/broadPeak/multiqc_data/"
},
{
"@type": "File",
"dateModified": "2020-09-10T12:26:59.191Z",
"name": "multiqc_report.html",
"@reverse": {
"hasPart": [
{
"@id": "results/multiqc/broadPeak/"
}
]
},
"@id": "results/multiqc/broadPeak/multiqc_report.html"
},
{
"@type": "Dataset",
"creator": {
"@id": "#db65dfb7-4867-400e-a12f-a1652d46a333"
},
"dateModified": "2020-09-10T12:27:01.599Z",
"hasPart": {
"@id": "results/pipeline_info/pipeline_dag.svg"
},
"name": "pipeline_info",
"@reverse": {
"hasPart": [
{
"@id": "results/"
}
]
},
"@id": "results/pipeline_info/"
},
{
"@type": [
"WorkflowSketch",
"File"
],
"about": {
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
},
"dateModified": "2020-09-10T12:27:01.755Z",
"encodingFormat": "image/svg+xml",
"name": "pipeline_dag.svg",
"@reverse": {
"hasPart": [
{
"@id": "results/pipeline_info/"
}
]
},
"@id": "results/pipeline_info/pipeline_dag.svg"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T11:57:13.996Z",
"hasPart": [
{
"@id": "results/trim_galore/fastqc/"
},
{
"@id": "results/trim_galore/logs/"
}
],
"name": "trim_galore",
"@reverse": {
"hasPart": [
{
"@id": "results/"
}
]
},
"@id": "results/trim_galore/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T11:58:55.705Z",
"hasPart": {
"@id": "results/trim_galore/fastqc/zips/"
},
"name": "fastqc",
"@reverse": {
"hasPart": [
{
"@id": "results/trim_galore/"
}
]
},
"@id": "results/trim_galore/fastqc/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T11:58:55.705Z",
"name": "zips",
"@reverse": {
"hasPart": [
{
"@id": "results/trim_galore/fastqc/"
}
]
},
"@id": "results/trim_galore/fastqc/zips/"
},
{
"@type": "Dataset",
"dateModified": "2020-09-10T11:58:55.705Z",
"name": "logs",
"@reverse": {
"hasPart": [
{
"@id": "results/trim_galore/"
}
]
},
"@id": "results/trim_galore/logs/"
},
{
"@type": "Person",
"name": "Phil Ewels",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#0164006f-bd58-4ebc-9a50-b8bd4ac3025c"
},
{
"@type": "CreativeWork",
"identifier": "https://spdx.org/licenses/CC0-1.0",
"name": "Creative Commons Zero v1.0 Universal",
"@reverse": {
"license": [
{
"@id": "./"
},
{
"@id": "chipseq_20200910.json"
},
{
"@id": "https://github.com/biocompute-objects/bco-ro-example-chipseq/archive/main.zip"
}
]
},
"@id": "https://spdx.org/licenses/CC0-1.0"
},
{
"@type": "Person",
"name": "Alexander Peltzer",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#556c747c-376a-4a85-82a1-9b99520d24fd"
},
{
"@type": "Person",
"name": "Winni Kretzschmar",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#64bd387d-60ad-4df8-804e-1f6b9ea72de5"
},
{
"@type": "Person",
"name": "Harshil Patel",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#714de175-aa77-47f1-9f99-6a4fba65530a"
},
{
"@type": "Person",
"name": "Drew Behrens",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#781b9b5a-dc06-4709-8f14-65ee08b8c543"
},
{
"@type": "Person",
"name": "Tiago Chedraoui Silva",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#93c23523-03b5-41dc-be4c-6a9a2e0e221d"
},
{
"@type": "Person",
"name": "mashehu",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#a58abf42-751d-49bd-a477-1d5065ac70c6"
},
{
"@type": "Person",
"name": "Chuan Wang",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#bfb876e7-e767-4209-ad66-e1e1379c249f"
},
{
"@type": "Person",
"name": "Nextflow 19.10.0",
"@reverse": {
"creator": [
{
"@id": "nextflow.log"
},
{
"@id": "results/"
},
{
"@id": "results/pipeline_info/"
}
]
},
"@id": "#db65dfb7-4867-400e-a12f-a1652d46a333"
},
{
"@type": "Person",
"name": "Rotholandus",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#e11af59b-8e24-4cc8-8f5e-cef411ab0823"
},
{
"@type": "Person",
"name": "Sofia Haglund",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#f262954b-a218-480d-8a01-0e0b1ca20ffc"
},
{
"@type": "Person",
"name": "Maxime Garcia",
"@reverse": {
"author": [
{
"@id": "https://raw.githubusercontent.com/nf-core/chipseq/1.2.1/main.nf"
}
]
},
"@id": "#f652b13e-0ba2-4394-a990-7304f54c7b9a"
},
{
"@type": "PropertyValue",
"name": "object_id",
"value": "dc308d7c-7949-446a-9c39-511b8ab40caf",
"@reverse": {
"identifier": [
{
"@id": "chipseq_20200910.json"
}
]
},
"@id": "urn:uuid:dc308d7c-7949-446a-9c39-511b8ab40caf"
}
]
}
RO-Crate for enabling a
large number of
data citations
Credit: Paolo Manghi
AGU Data Citation Workshop
https://doi.org/10.5281/zenodo.4916734
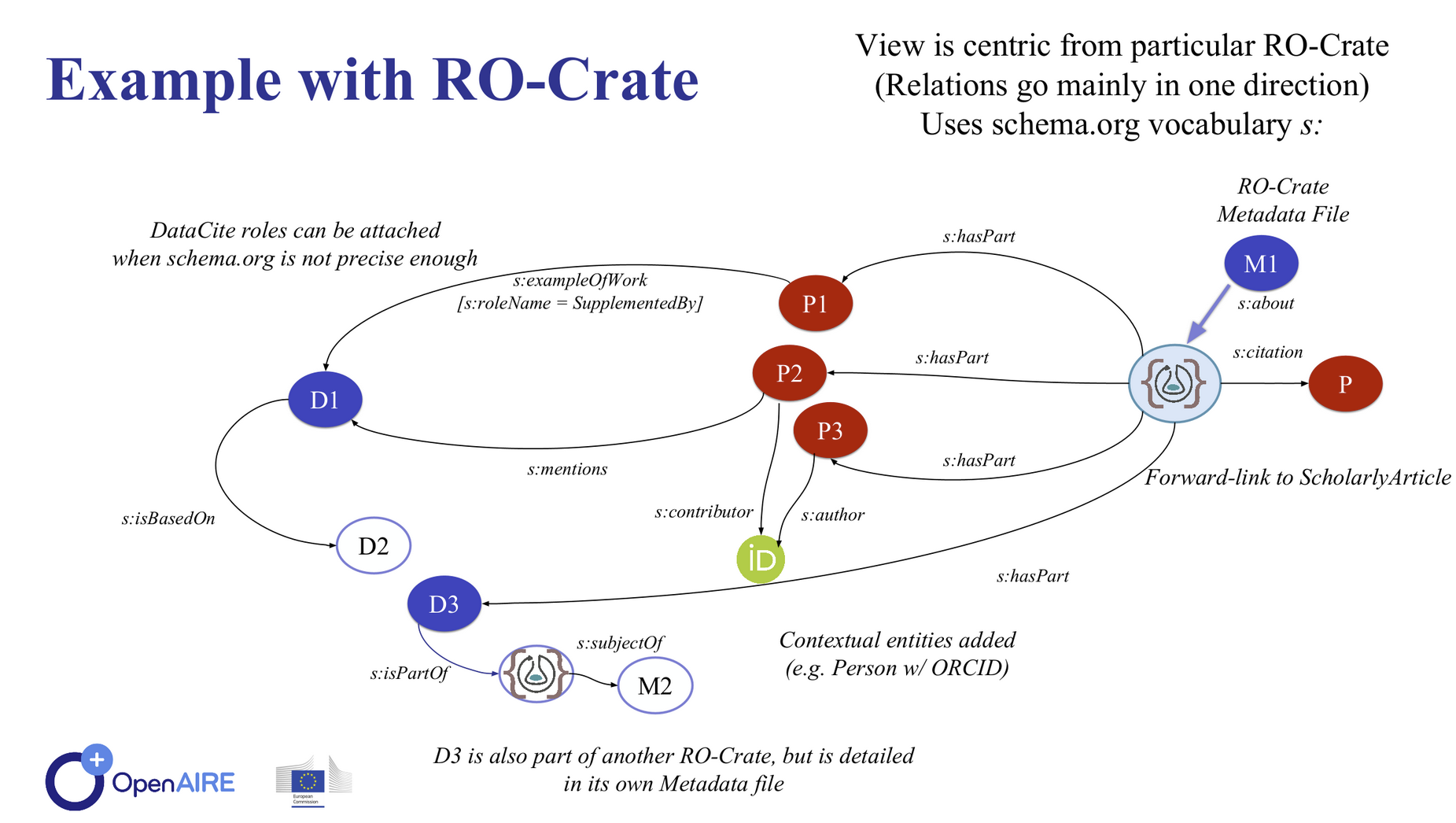
RO-Crate as aggregation:
data citation reliquary
4-dimensional RO-Crates?

Credit: Oscar Corcho, Carole Goble
https://doi.org/10.5281/zenodo.4913285
RO-Crate profiles
Credit: Carole Goble
Dataverse Community Meeting 2021
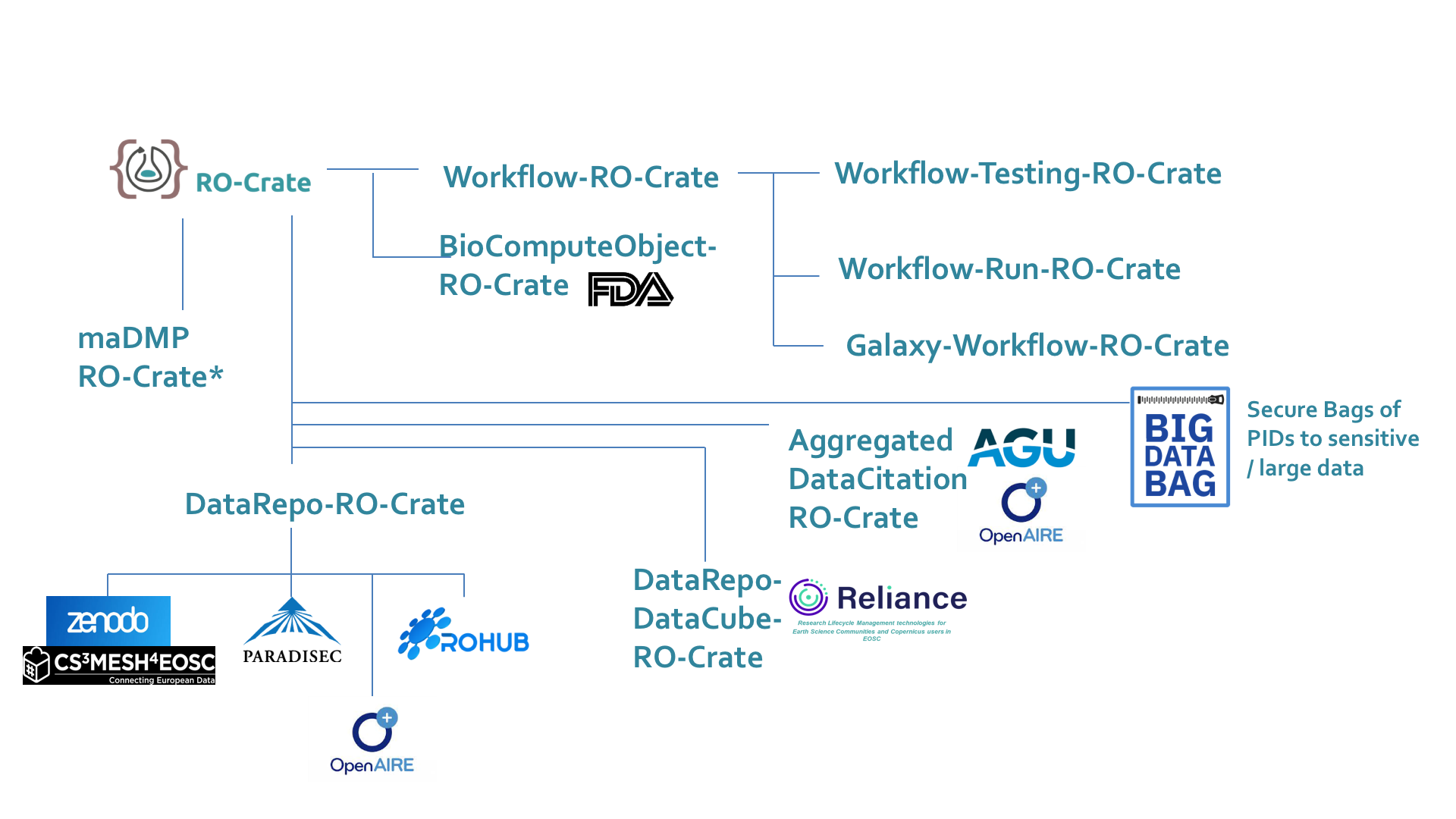
FAIR Digital Objects
…with RO-Crate as metadata object
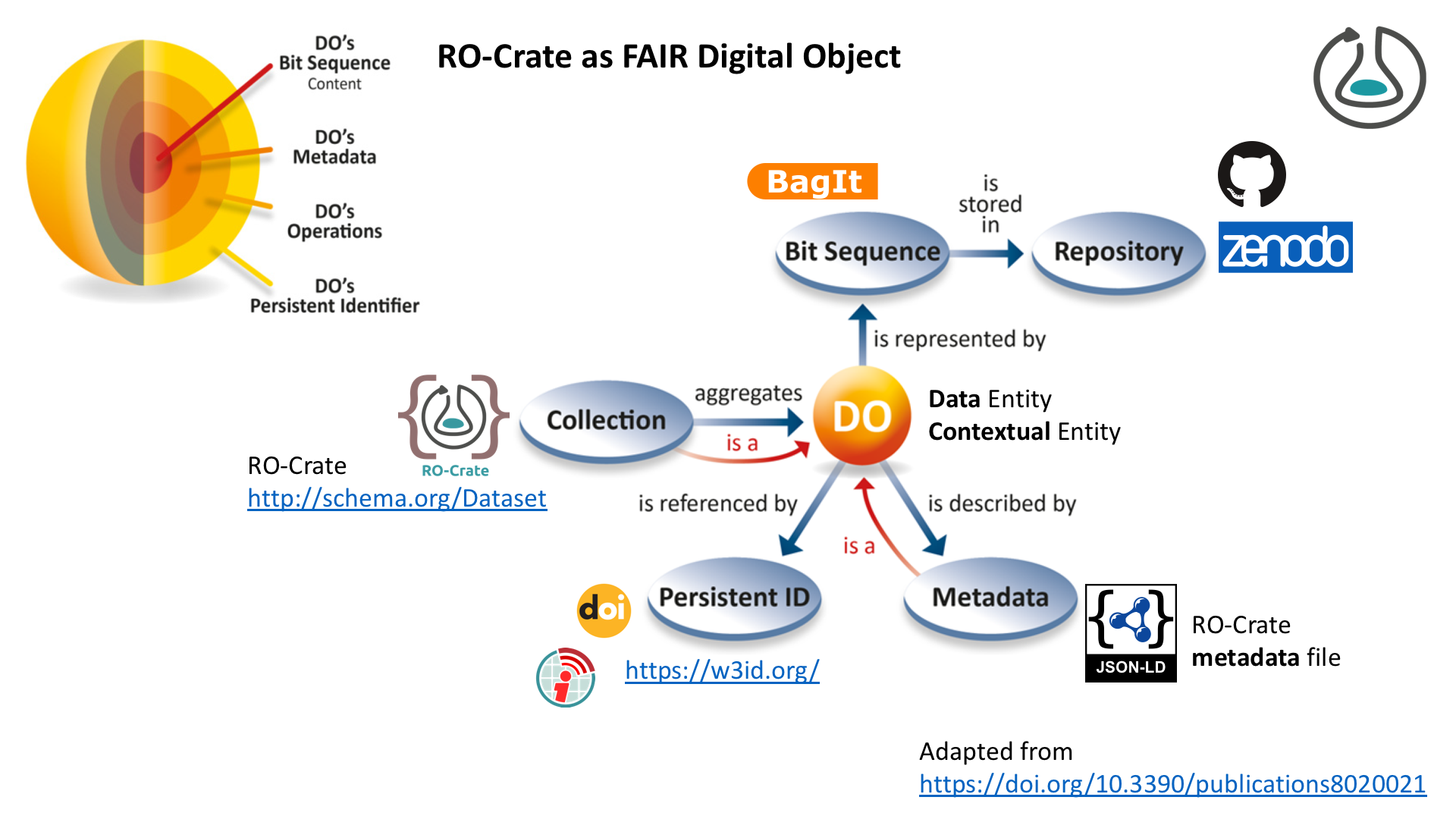
RO-Crate as FAIR Digital Object (FDO)
+ FAIR Signposting
Credit:
Herbert van de Sompel
FAIR Signposting: A KISS Approach to a Burning Issue
FAIR Signposting
Credit:
Herbert van de Sompel
FAIR Signposting: A KISS Approach to a Burning Issue
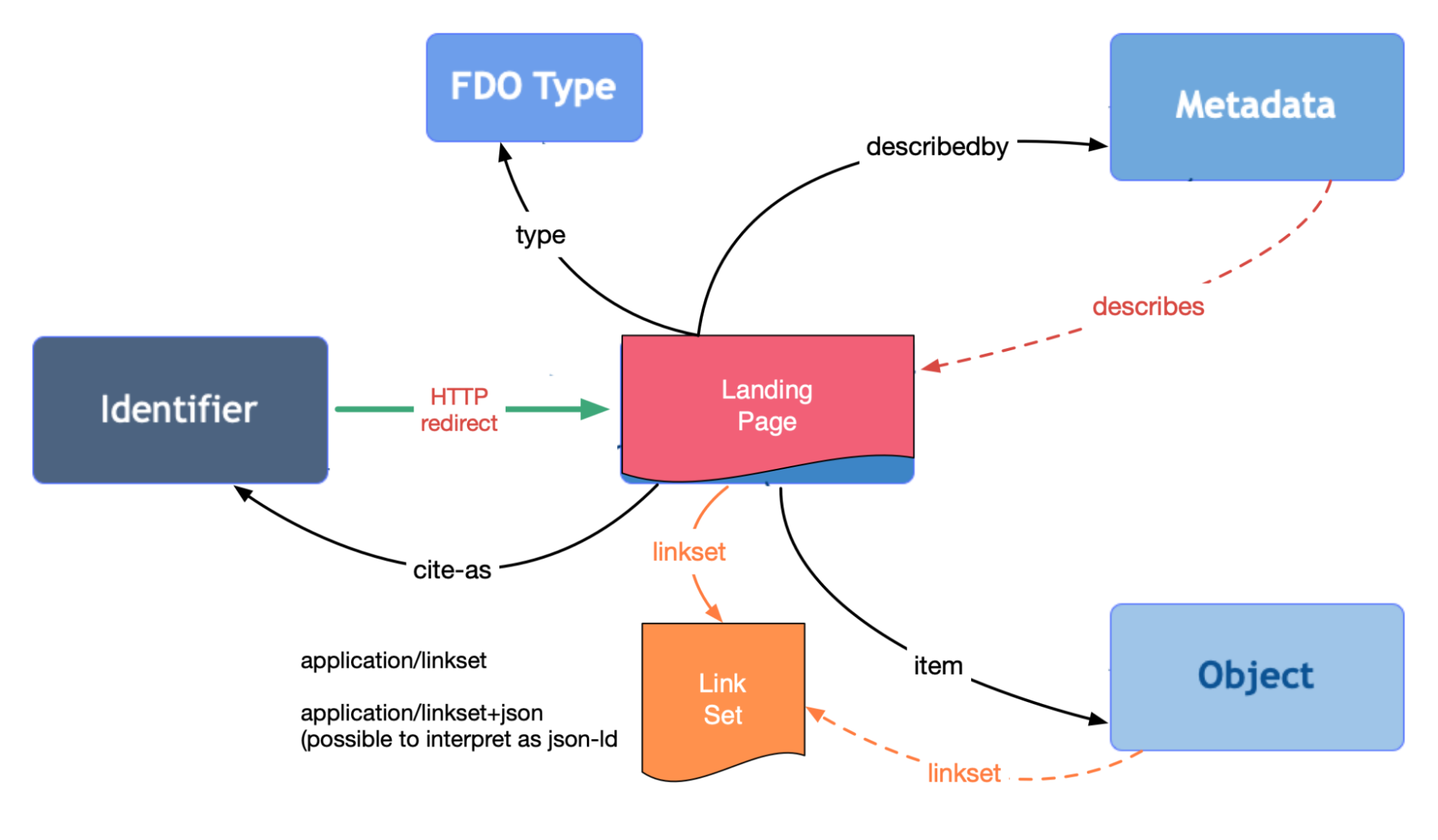
FAIR Signposting
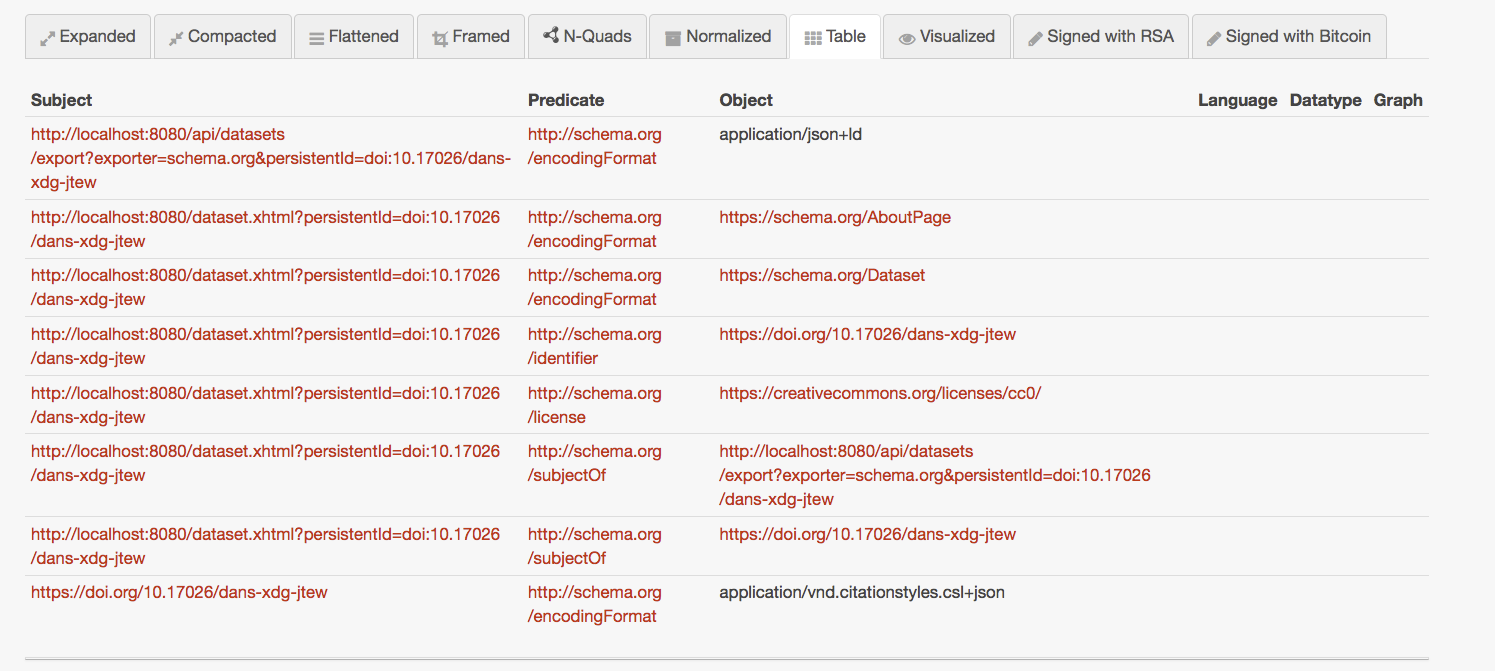
HEAD https://doi.org/10.17026/dans-xdg-jtew HTTP/2
Accept: */*
HTTP/2 302
date: Tue, 29 Jun 2021 15:02:06 GMT
content-type: text/html;charset=utf-8
vary: Accept
location: https://easy.dans.knaw.nl/ui/datasets/id/easy-dataset:32697
expires: Tue, 29 Jun 2021 15:28:25 GMT
…

HEAD https://doi.org/10.17026/dans-xdg-jtew HTTP/2
Accept: */*
HTTP/2 302
date: Tue, 29 Jun 2021 15:02:06 GMT
content-type: text/html;charset=utf-8
vary: Accept
location: https://easy.dans.knaw.nl/ui/datasets/id/easy-dataset:32697
expires: Tue, 29 Jun 2021 15:28:25 GMT
…
HEAD https://easy.dans.knaw.nl/ui/datasets/id/easy-dataset:32697 HTTP/1.1
Connection: close
HTTP/1.1 200 OK
Date: Tue, 29 Jun 2021 15:02:06 GMT
Cache-Control: no-cache, max-age=0, must-revalidate
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="cite-as"
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="describedby" ; type="application/vnd.datacite.datacite+xml"
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="describedby" ; type="application/vnd.citationstyles.csl+json"
Link: <http://www.persistent-identifier.nl?identifier=urn%3Anbn%3Anl%3Aui%3A13-k7v-xhk> ; rel="cite-as"
Link: <https://easy.dans.knaw.nl/ui/resources/easy/export?sid=easy-dataset%3A32697&format=XML> ;
rel="describedby" ; type="application/xml" ; profile="https://easy.dans.knaw.nl/easy/easymetadata/",
<https://easy.dans.knaw.nl/ui/resources/easy/export?sid=easy-dataset%3A32697&format=CSV> ;
rel="describedby" ; type="txt/csv"
Content-Type: text/html;charset=UTF-8
Content-Language: en-US
Strict-Transport-Security: max-age=31536000; includeSubDomainsHEAD https://easy.dans.knaw.nl/ui/datasets/id/easy-dataset:32697 HTTP/1.1
Connection: close
HTTP/1.1 200 OK
Date: Tue, 29 Jun 2021 15:02:06 GMT
Cache-Control: no-cache, max-age=0, must-revalidate
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="cite-as"
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="describedby" ; type="application/vnd.datacite.datacite+xml"
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="describedby" ; type="application/vnd.citationstyles.csl+json"
Link: <http://www.persistent-identifier.nl?identifier=urn%3Anbn%3Anl%3Aui%3A13-k7v-xhk> ; rel="cite-as"
Link: <https://easy.dans.knaw.nl/ui/resources/easy/export?sid=easy-dataset%3A32697&format=XML> ;
rel="describedby"; type="application/xml" ; profile="https://easy.dans.knaw.nl/easy/easymetadata/",
<https://easy.dans.knaw.nl/ui/resources/easy/export?sid=easy-dataset%3A32697&format=CSV> ;
rel="describedby"; type="txt/csv"
Link: <http://example.com/api/datasets/export?exporter=schema.org&persistentId=doi:10.17026/dans-xdg-jtew>;
rel="describedby"; type="application/json+ld",
Link: <https://schema.org/AboutPage>; rel="type",
<https://schema.org/Dataset>; rel="type",
Link: https://creativecommons.org/licenses/cc0/;rel="license"
Link: <http://example.com/api/datasets/:persistentId/versions/1.0/linkset?persistentId=doi:10.17026/dans-xdg-jtew> ;
rel="linkset"; type="application/linkset+json"
Content-Type: text/html;charset=UTF-8
Content-Language: en-US
Strict-Transport-Security: max-age=31536000; includeSubDomainsapplication/linkset+json
https://datatracker.ietf.org/doc/html/draft-ietf-httpapi-linkset-02
HEAD https://easy.dans.knaw.nl/ui/datasets/id/easy-dataset:32697 HTTP/1.1
Connection: close
HTTP/1.1 200 OK
Date: Tue, 29 Jun 2021 15:02:06 GMT
Cache-Control: no-cache, max-age=0, must-revalidate
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="cite-as"
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="describedby" ; type="application/vnd.datacite.datacite+xml"
Link: <https://doi.org/10.17026/dans-xdg-jtew> ; rel="describedby" ; type="application/vnd.citationstyles.csl+json"
Link: <http://www.persistent-identifier.nl?identifier=urn%3Anbn%3Anl%3Aui%3A13-k7v-xhk> ; rel="cite-as"
Link: <https://easy.dans.knaw.nl/ui/resources/easy/export?sid=easy-dataset%3A32697&format=XML> ;
rel="describedby"; type="application/xml" ; profile="https://easy.dans.knaw.nl/easy/easymetadata/",
<https://easy.dans.knaw.nl/ui/resources/easy/export?sid=easy-dataset%3A32697&format=CSV> ;
rel="describedby"; type="txt/csv"
Link: <http://example.com/api/datasets/export?exporter=schema.org&persistentId=doi:10.17026/dans-xdg-jtew>;
rel="describedby"; type="application/json+ld",
Link: <https://schema.org/AboutPage>; rel="type",
<https://schema.org/Dataset>; rel="type",
Link: https://creativecommons.org/licenses/cc0/;rel="license"
Link: <http://example.com/api/datasets/:persistentId/versions/1.0/linkset?persistentId=doi:10.17026/dans-xdg-jtew> ;
rel="linkset"; type="application/linkset+json"
Content-Type: text/html;charset=UTF-8
Content-Language: en-US
Strict-Transport-Security: max-age=31536000; includeSubDomains{
"linkset": [
{
"anchor": "http://localhost:8080/dataset.xhtml?persistentId=doi:10.17026/dans-xdg-jtew",
"cite-as": [
{
"href": "https://doi.org/10.17026/dans-xdg-jtew"
}
],
"type": [
{
"href": "https://schema.org/AboutPage"
},
{
"href": "https://schema.org/Dataset"
}
],
"license": {
"href": "https://creativecommons.org/licenses/cc0/"
},
"describedby": [
{
"href": "https://doi.org/10.17026/dans-xdg-jtew",
"type": "application/vnd.citationstyles.csl+json"
},
{
"href": "http://localhost:8080/api/datasets/export?exporter=schema.org&persistentId=doi:10.17026/dans-xdg-jtew",
"type": "application/json+ld"
}
],
"item": []
}
]
}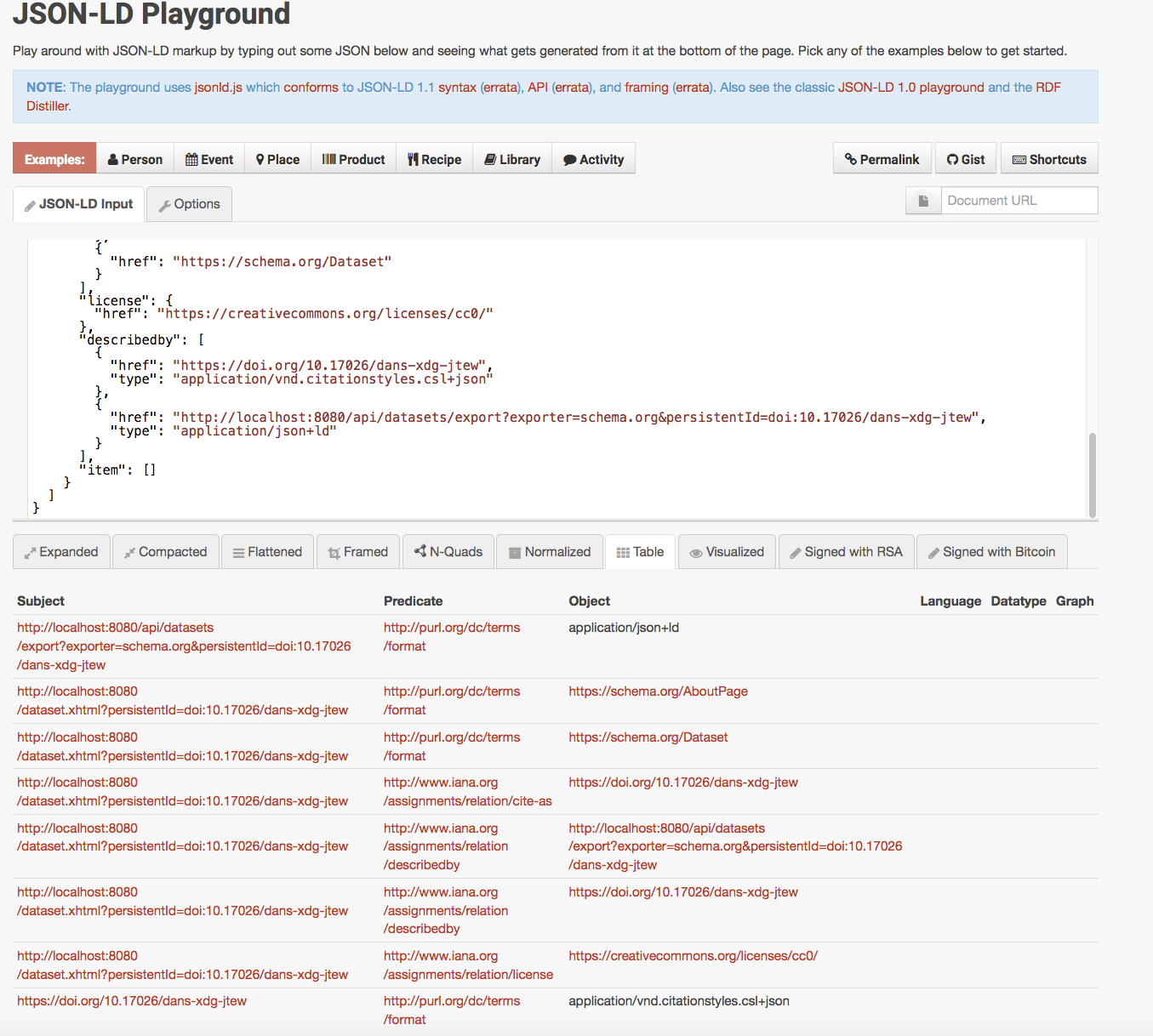
Interpreting application/linkset+json as JSON-LD
… or mapped to schema.org
FDO for Workflow RO-Crate



302 Found Location: https://workflowhub.eu/workflows/29?version=2
Accept: text/html
Resolving an RO-Crate with content-negotiation


ComputationalWorkflow




302 Found Location: https://workflowhub.eu/workflows/29?version=2
Accept: text/html

Accept: application/zip



Resolving an RO-Crate with content-negotiation







302 Found Location: https://workflowhub.eu/workflows/29?version=2
Accept: text/html
Accept: application/ld+json; profile=https://w3id.org/ro/crate



Accept: application/zip





Resolving an RO-Crate with content-negotiation





302 Found Location: https://workflowhub.eu/workflows/29?version=2
Accept: application/ld+json; profile=https://w3id.org/ro/crate




Downside: Indirection to find core metadata


author
@type

ComputationalWorkflow

license
hasPart
isBasedOn
Parse JSON, find the right node


HEAD https://doi.org/10.48546/workflowhub.workflow.29.2 302 Found Location: https://workflowhub.eu/workflows/29?version=2
HEAD https://workflowhub.eu/workflows/29?version=2 200 OK Link: <https://doi.org/10.48546/workflowhub.workflow.29.2>;rel=cite-as Link: <https://workflowhub.eu/workflows/29/ro_crate?version=2>;rel=describedby Link: <https://orcid.org/0000-0003-0513-0288>;rel=author …



Resolving an RO-Crate with FAIR Signposting


rel=author
rel=type

ComputationalWorkflow

rel=license
rel=item
rel=describedby;
type="application/ld+json;profile=https://w3id.org/ro/crate"
rel=cite-as

rel=item;
type="application/zip"
More FDOs?
More workflow FDOs?
Canonical workflow (e.g. BioBB)
Workflow entry (e.g. WorkflowHub)
Workflow definition (e.g. Galaxy file)
Example run of workflow (e.g. CWLProv, BCO)
Workflow visualizations (e.g. CWL Viewer)
Tool definitions used by workflow step
Container image run from definition
(Recipe for container image)
Software package(s) installed in container
Entry in software registry (bio.tools, RRID, ASCL)
Software citations (e.g. JoSS)
Lessons learnt
Lessons Learnt
- Workflows are hetereogeneous
- Need to unify core metadata and extract structure
--> derived resources - Workflows may have many sub-components
- Need to unify core metadata and extract structure
- Workflows may already have a Web or Git presence
- … or may be uploaded from disk
- --> WorkflowHub as DOI provider
- Workflows involve existing digital objects 'out of our control'
- Tools, containers, GitHub Repos
- Tools and software are heterogeneous compound objects
- Workflow runs show (all) the details
--> need for explanation and context
- Paul Walk https://orcid.org/0000-0003-1541-5631
- brandon whitehead https://orcid.org/0000-0002-0337-8610
- Mark Wilkinson https://orcid.org/0000-0001-6960-357X
- Paul Groth https://orcid.org/0000-0003-0183-6910
- Erich Bremer https://orcid.org/0000-0003-0223-1059
- LJ Garcia Castro https://orcid.org/0000-0003-3986-0510
- Karl Sebby https://orcid.org/0000-0001-6022-9825
- Alexander Kanitz https://orcid.org/0000-0002-3468-0652
- Ana Trisovic https://orcid.org/0000-0003-1991-0533
- Gavin Kennedy https://orcid.org/0000-0003-3910-0474
- Mark Graves https://orcid.org/0000-0003-3486-8193
- Jasper Koehorst https://orcid.org/0000-0001-8172-8981
- Simone Leo https://orcid.org/0000-0001-8271-5429
- Marc Portier https://orcid.org/0000-0002-9648-6484
- Paul Brack https://orcid.org/0000-0002-5432-2748
- Milan Ojsteršek https://orcid.org/0000-0003-1743-8300
- Bert Droesbeke https://orcid.org/0000-0003-0522-5674
- Chenxu Niu https://orcid.org/0000-0002-2142-1731
- Kosuke Tanabe https://orcid.org/0000-0002-9986-7223
- Tomasz Miksa https://orcid.org/0000-0002-4929-7875
- Marco La Rosa https://orcid.org/0000-0001-5383-6993
- Cedric Decruw https://github.com/cedricdcc
- Andreas Czerniak https://orcid.org/0000-0003-3883-4169
- Jeremy Jay https://orcid.org/0000-0002-5761-7533
- Sergio Serra https://orcid.org/0000-0002-0792-8157
RO-Crate Community
- Peter Sefton https://orcid.org/0000-0002-3545-944X (co-chair)
- Stian Soiland-Reyes https://orcid.org/0000-0001-9842-9718 (co-chair)
- Eoghan Ó Carragáin https://orcid.org/0000-0001-8131-2150 (emeritus chair)
- Oscar Corcho https://orcid.org/0000-0002-9260-0753
- Daniel Garijo https://orcid.org/0000-0003-0454-7145
- Raul Palma https://orcid.org/0000-0003-4289-4922
- Frederik Coppens https://orcid.org/0000-0001-6565-5145
- Carole Goble https://orcid.org/0000-0003-1219-2137
- José María Fernández https://orcid.org/0000-0002-4806-5140
- Kyle Chard https://orcid.org/0000-0002-7370-4805
- Jose Manuel Gomez-Perez https://orcid.org/0000-0002-5491-6431
- Michael R Crusoe https://orcid.org/0000-0002-2961-9670
- Ignacio Eguinoa https://orcid.org/0000-0002-6190-122X
- Nick Juty https://orcid.org/0000-0002-2036-8350
- Kristi Holmes https://orcid.org/0000-0001-8420-5254
- Jason A. Clark https://orcid.org/0000-0002-3588-6257
- Salvador Capella-Gutierrez https://orcid.org/0000-0002-0309-604X
- Alasdair J. G. Gray https://orcid.org/0000-0002-5711-4872
- Stuart Owen https://orcid.org/0000-0003-2130-0865
- Alan R Williams https://orcid.org/0000-0003-3156-2105
- Giacomo Tartari https://orcid.org/0000-0003-1130-2154
- Finn Bacall https://orcid.org/0000-0002-0048-3300
- Thomas Thelen https://orcid.org/0000-0002-1756-2128
- Hervé Ménager https://orcid.org/0000-0002-7552-1009
- Laura Rodríguez-Navas https://orcid.org/0000-0003-4929-1219
The RO-Crate Community is open for anyone to join us!