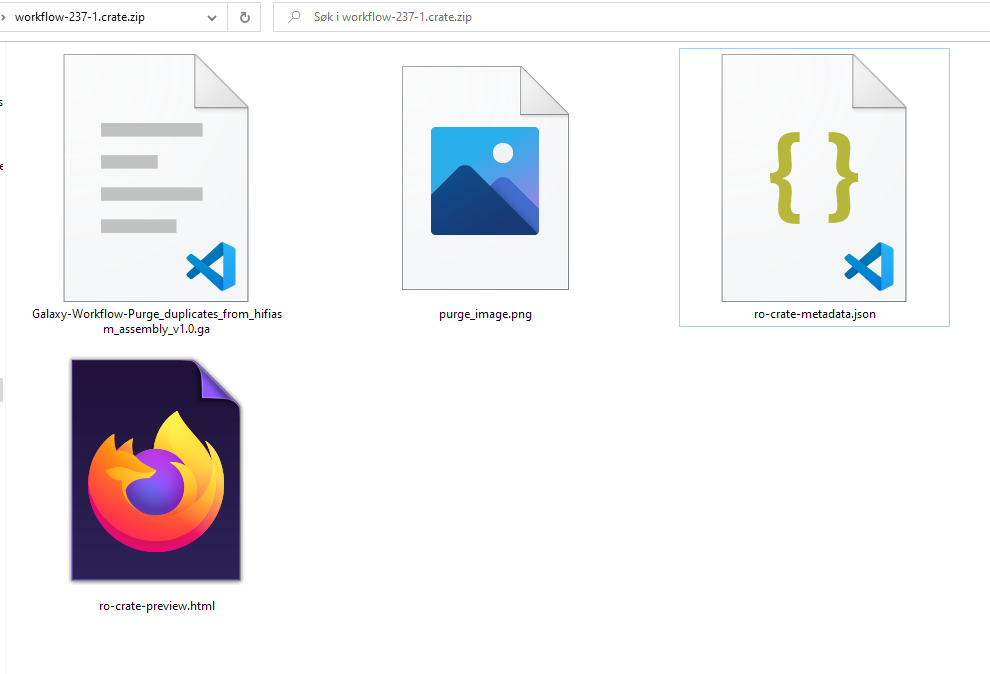
RO-Crate
A brief "crash course"
Stian Soiland-Reyes
eScience lab, The University of Manchester
INDElab, University of Amsterdam
ELIXIR Data-Interoperability F2F
2021-11-23
This work is licensed under a
Creative Commons Attribution 4.0 International License.









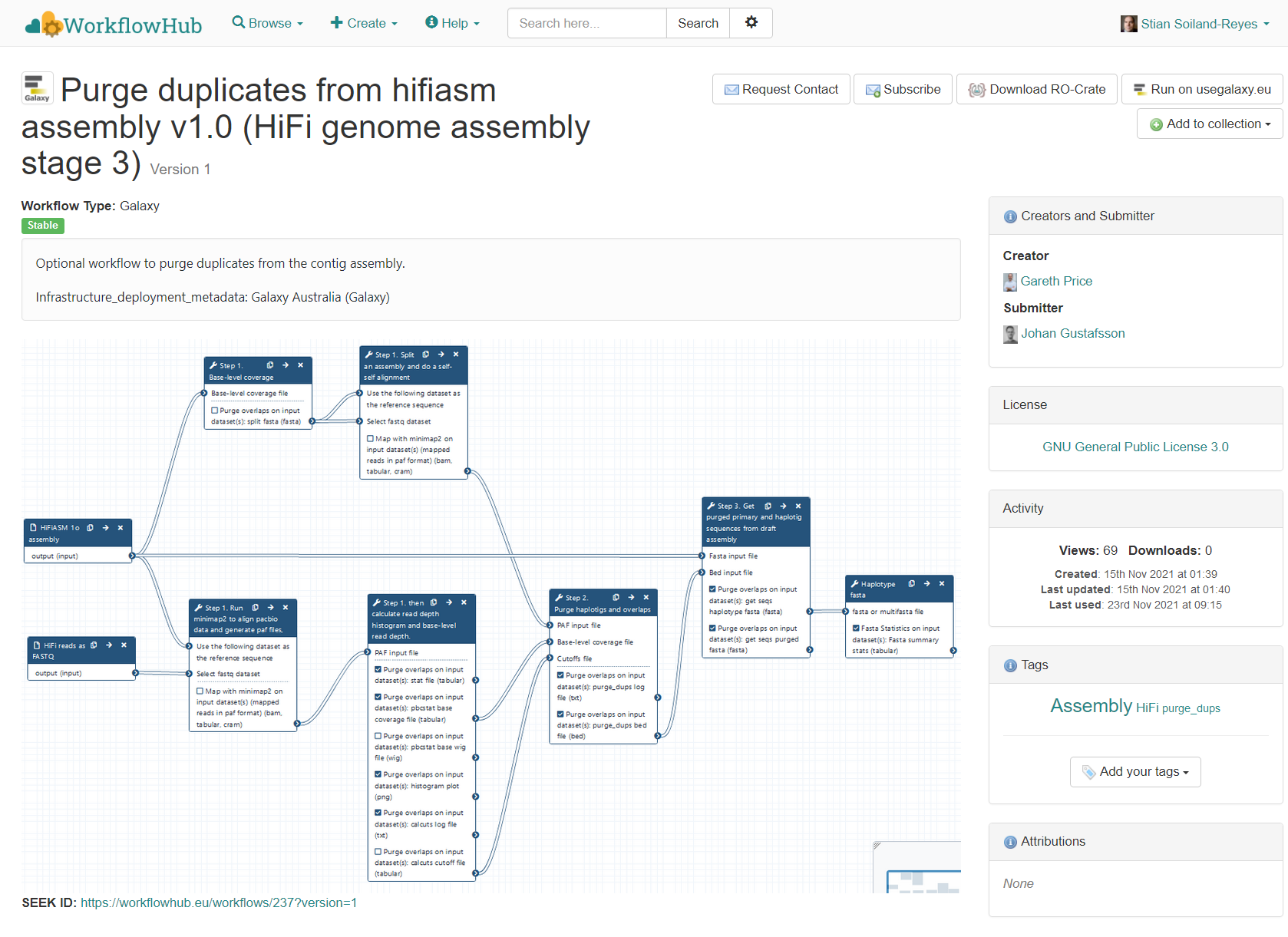

{
"@context": "https://w3id.org/ro/crate/1.1/context",
"@graph": [
{
"@id": "ro-crate-metadata.json",
"@type": "CreativeWork",
"about": {
"@id": "./"
},
"conformsTo": [
{
"@id": "https://w3id.org/ro/crate/1.1"
},
{
"@id": "https://about.workflowhub.eu/Workflow-RO-Crate/"
}
]
},
{
"@id": "ro-crate-preview.html",
"@type": "CreativeWork",
"about": {
"@id": "./"
}
},
{
"@id": "./",
"@type": "Dataset",
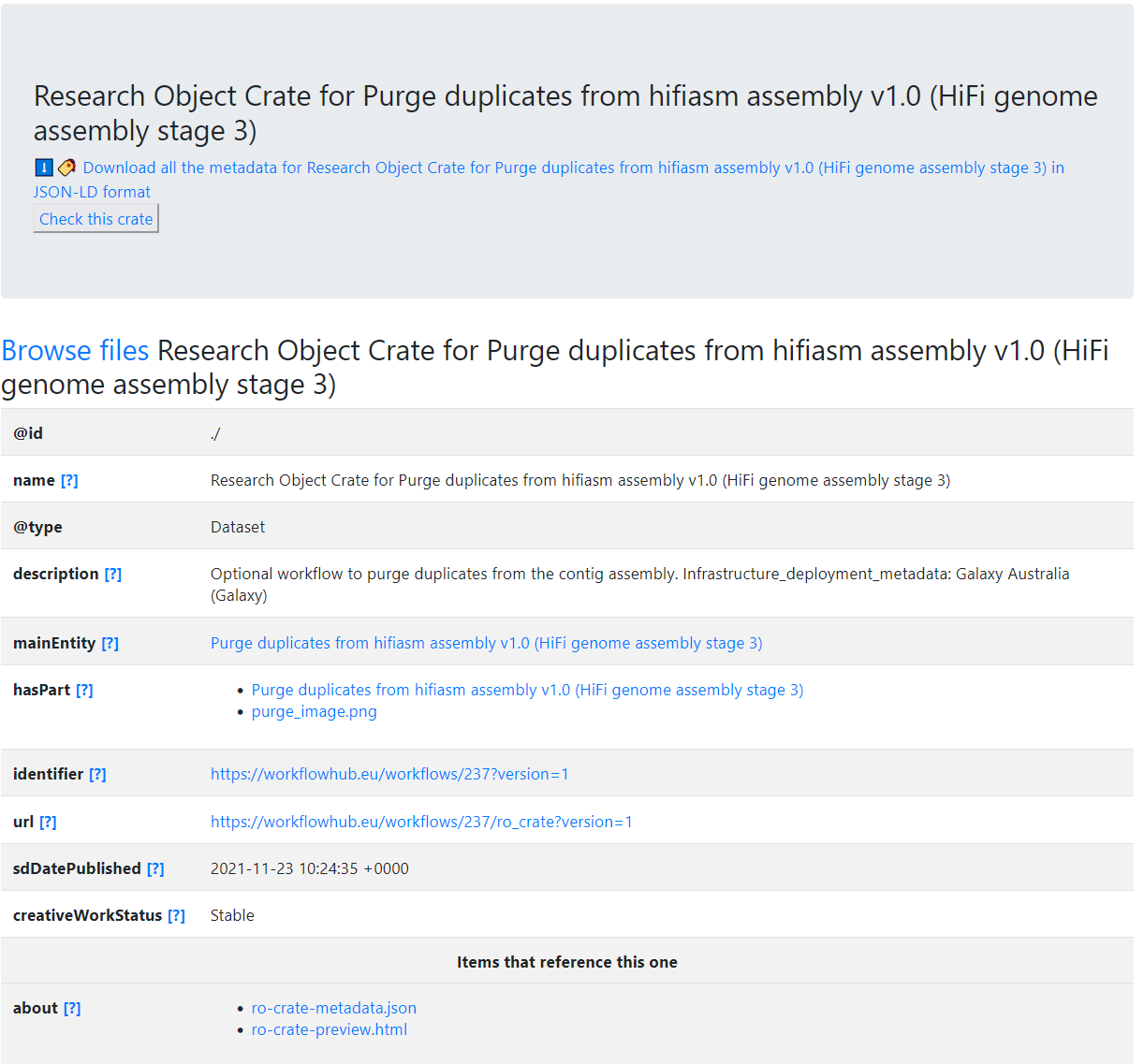
"mainEntity": {
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga"
},
"hasPart": [
{
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga"
},
{
"@id": "purge_image.png"
}
],
"identifier": "https://workflowhub.eu/workflows/237?version=1",
"url": "https://workflowhub.eu/workflows/237/ro_crate?version=1",
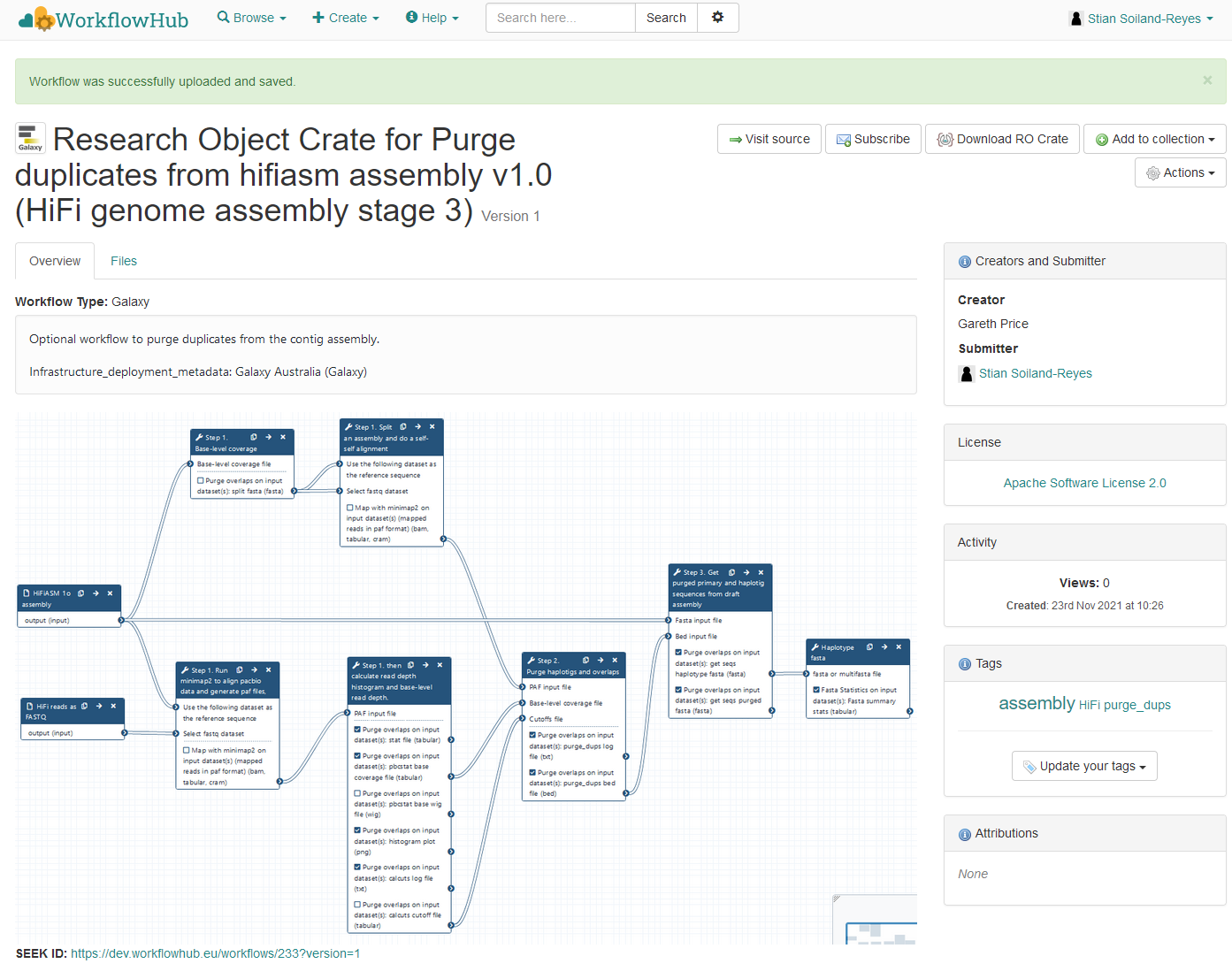
"name": "Research Object Crate for Purge duplicates from hifiasm assembly v1.0 (HiFi genome assembly stage 3)",
"description": "Optional workflow to purge duplicates from the contig assembly.\r\n\r\nInfrastructure_deployment_metadata: Galaxy Australia (Galaxy)",
"sdDatePublished": "2021-11-23 10:44:32 +0000",
"creativeWorkStatus": "Stable"
},
{
"@id": "Galaxy-Workflow-Purge_duplicates_from_hifiasm_assembly_v1.0.ga",
"@type": [
"File",
"SoftwareSourceCode",
"ComputationalWorkflow"
],
"programmingLanguage": {
"@id": "#galaxy"
},
"image": {
"@id": "purge_image.png"
},
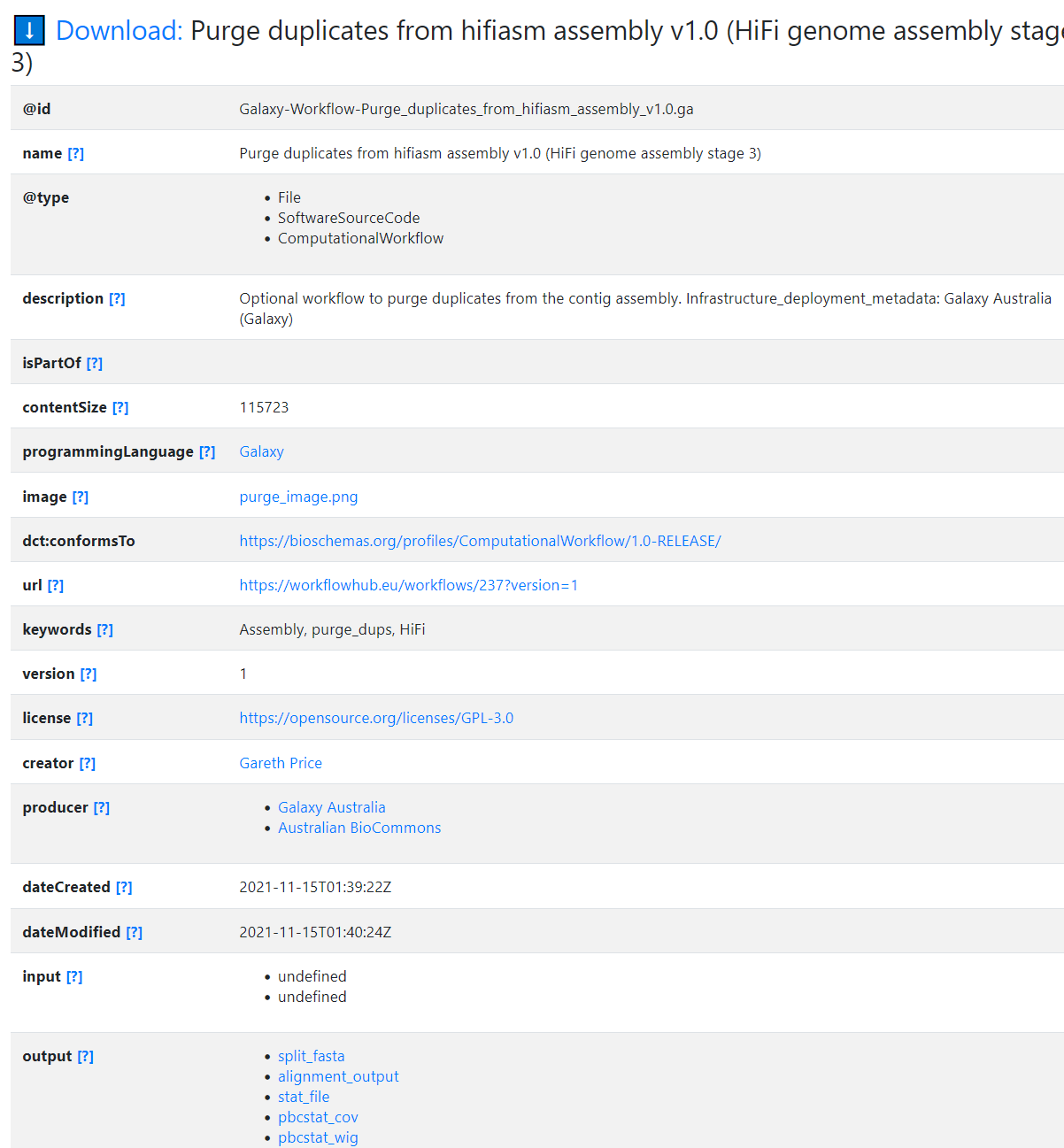
"contentSize": 115723,
"dct:conformsTo": "https://bioschemas.org/profiles/ComputationalWorkflow/1.0-RELEASE/",
"description": "Optional workflow to purge duplicates from the contig assembly.\r\n\r\nInfrastructure_deployment_metadata: Galaxy Australia (Galaxy)",
"name": "Purge duplicates from hifiasm assembly v1.0 (HiFi genome assembly stage 3)",
"url": "https://workflowhub.eu/workflows/237?version=1",
"keywords": "Assembly, purge_dups, HiFi",
"version": 1,
"license": "https://opensource.org/licenses/GPL-3.0",
"creator": {
"@id": "https://workflowhub.eu/people/139"
},
"producer": [
{
"@id": "https://workflowhub.eu/projects/54"
},
{
"@id": "https://workflowhub.eu/projects/30"
}
],
"dateCreated": "2021-11-15T01:39:22Z",
"dateModified": "2021-11-15T01:40:24Z",
"isPartOf": [
],
"input": [
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFiASM 1o assembly"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFi reads as FASTQ"
}
],
"output": [
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-split_fasta"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-alignment_output"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stat_file"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_cov"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_wig"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-hist"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_log"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_tab"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_log"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_bed"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_hap"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_purged"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stats"
}
],
"sdPublisher": {
"@id": "http://about.workflowhub.eu"
}
},
{
"@id": "purge_image.png",
"@type": [
"File",
"ImageObject",
"WorkflowSketch"
],
"contentSize": 118673
},
{
"@id": "https://about.workflowhub.eu/Workflow-RO-Crate/",
"@type": "CreativeWork",
"name": "Workflow RO-Crate Profile",
"version": "0.2.0"
},
{
"@id": "https://workflowhub.eu/people/139",
"@type": "Person",
"name": "Gareth Price"
},
{
"@id": "https://workflowhub.eu/projects/54",
"@type": [
"Project",
"Organization"
],
"name": "Galaxy Australia"
},
{
"@id": "https://workflowhub.eu/projects/30",
"@type": [
"Project",
"Organization"
],
"name": "Australian BioCommons"
},
{
"@id": "#galaxy",
"@type": "ComputerLanguage",
"name": "Galaxy",
"identifier": {
"@id": "https://galaxyproject.org/"
},
"url": {
"@id": "https://galaxyproject.org/"
}
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFiASM%201o%20assembly",
"@type": "FormalParameter",
"name": "HiFiASM 1o assembly",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-inputs-HiFi%20reads%20as%20FASTQ",
"@type": "FormalParameter",
"name": "HiFi reads as FASTQ",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-split_fasta",
"@type": "FormalParameter",
"name": "split_fasta",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-alignment_output",
"@type": "FormalParameter",
"name": "alignment_output",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stat_file",
"@type": "FormalParameter",
"name": "stat_file",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_cov",
"@type": "FormalParameter",
"name": "pbcstat_cov",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-pbcstat_wig",
"@type": "FormalParameter",
"name": "pbcstat_wig",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-hist",
"@type": "FormalParameter",
"name": "hist",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_log",
"@type": "FormalParameter",
"name": "calcuts_log",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-calcuts_tab",
"@type": "FormalParameter",
"name": "calcuts_tab",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_log",
"@type": "FormalParameter",
"name": "purge_dups_log",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-purge_dups_bed",
"@type": "FormalParameter",
"name": "purge_dups_bed",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_hap",
"@type": "FormalParameter",
"name": "get_seqs_hap",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-get_seqs_purged",
"@type": "FormalParameter",
"name": "get_seqs_purged",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "#purge_duplicates_from_hifiasm_assembly_v1_0__hifi_genome_assembly_stage_3_-outputs-stats",
"@type": "FormalParameter",
"name": "stats",
"dct:conformsTo": "https://bioschemas.org/profiles/FormalParameter/1.0-RELEASE/"
},
{
"@id": "http://about.workflowhub.eu",
"@type": "Organization",
"name": "WorkflowHub",
"url": "http://about.workflowhub.eu"
}
]
}
ro-crate-metadata.json


Text
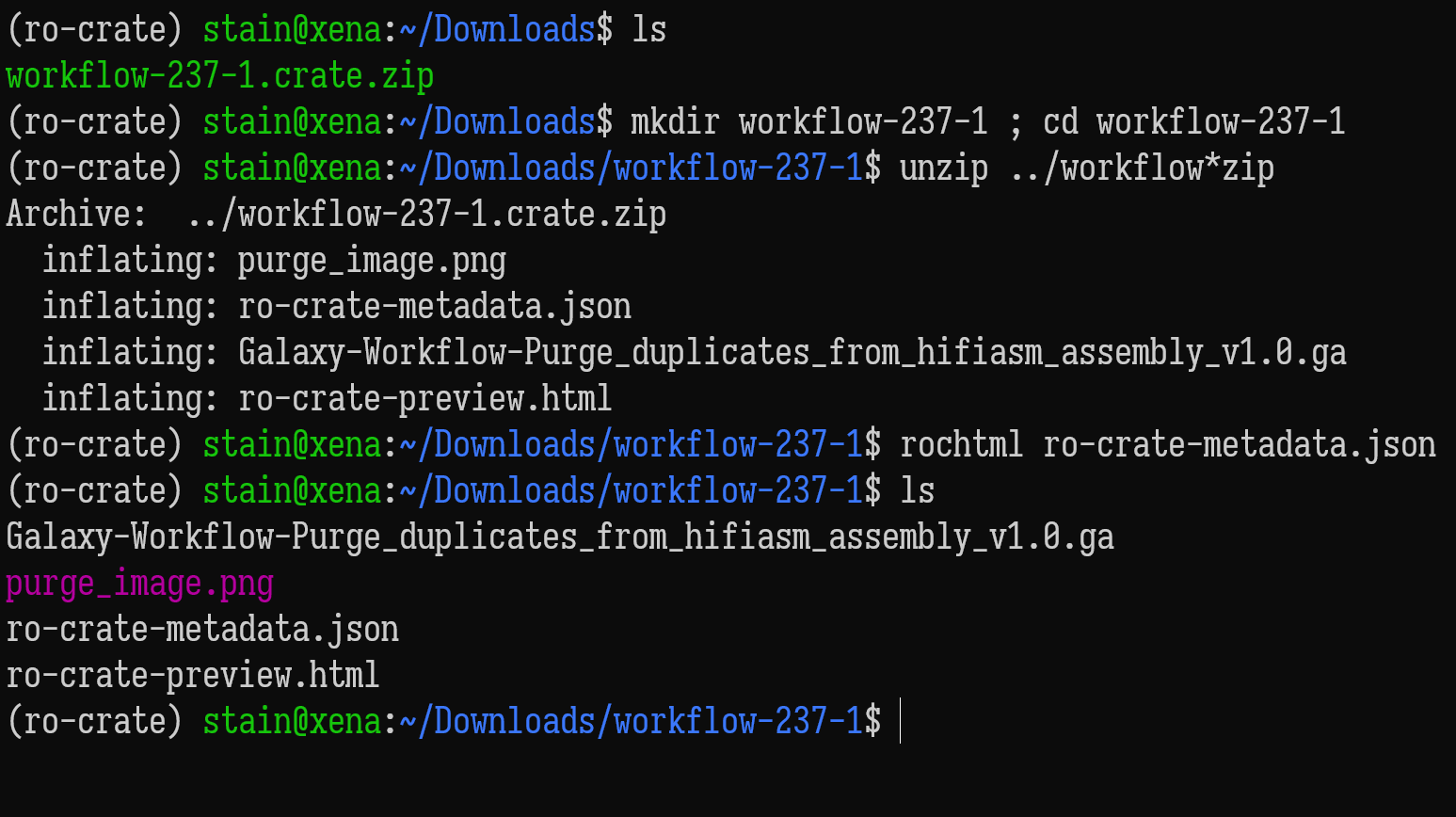
Generate a HTML preview from JSON-LD





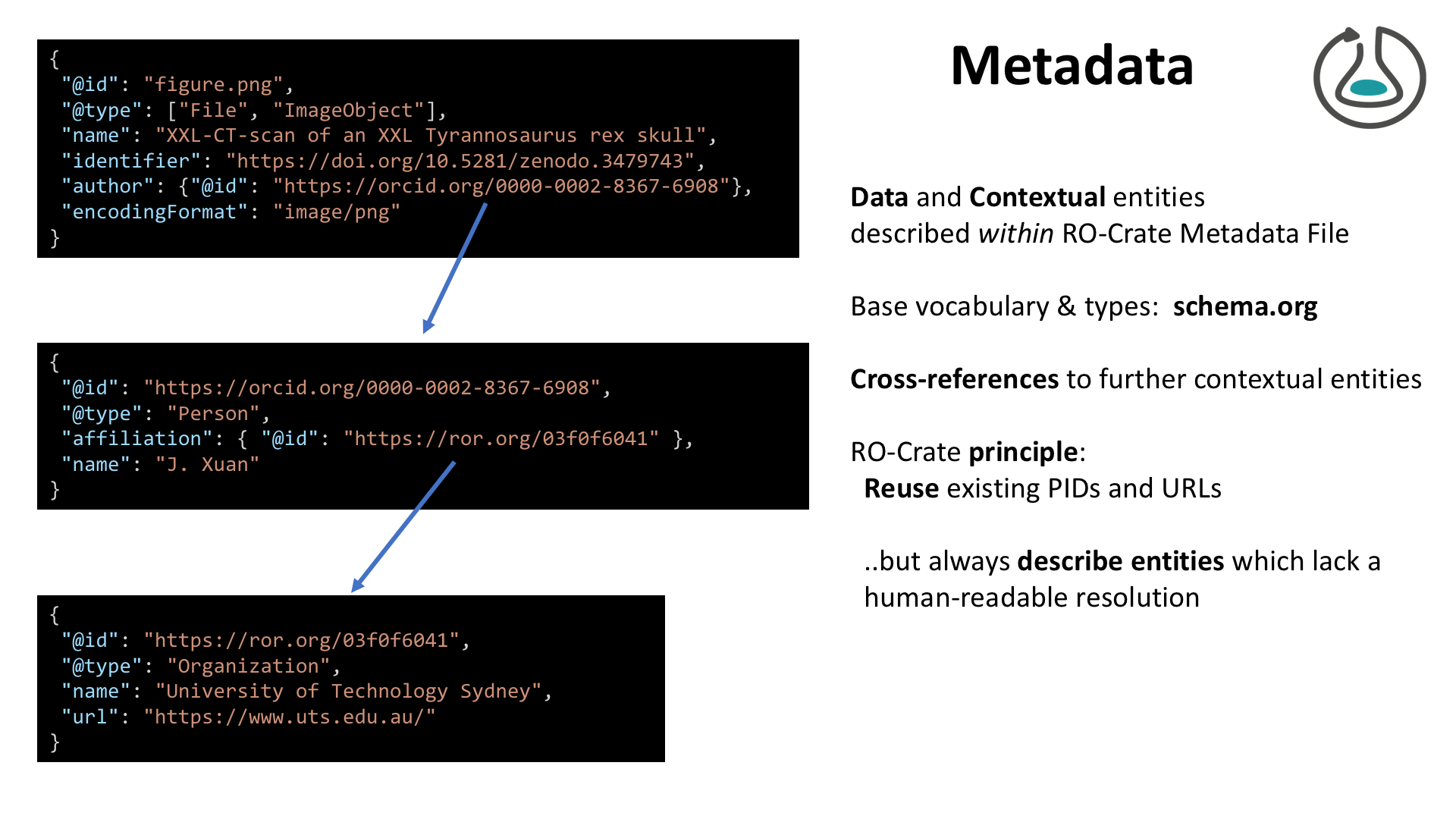
Techie deep-dive!
Warning: JSON ahead















Simone Leo; Stian Soiland-Reyes; Ignacio Eguinoa;
Bert Droesbeke; Alban Gaignard; Laura Rodríguez Navas
Credit: Marco La Rosa, Peter Sefton
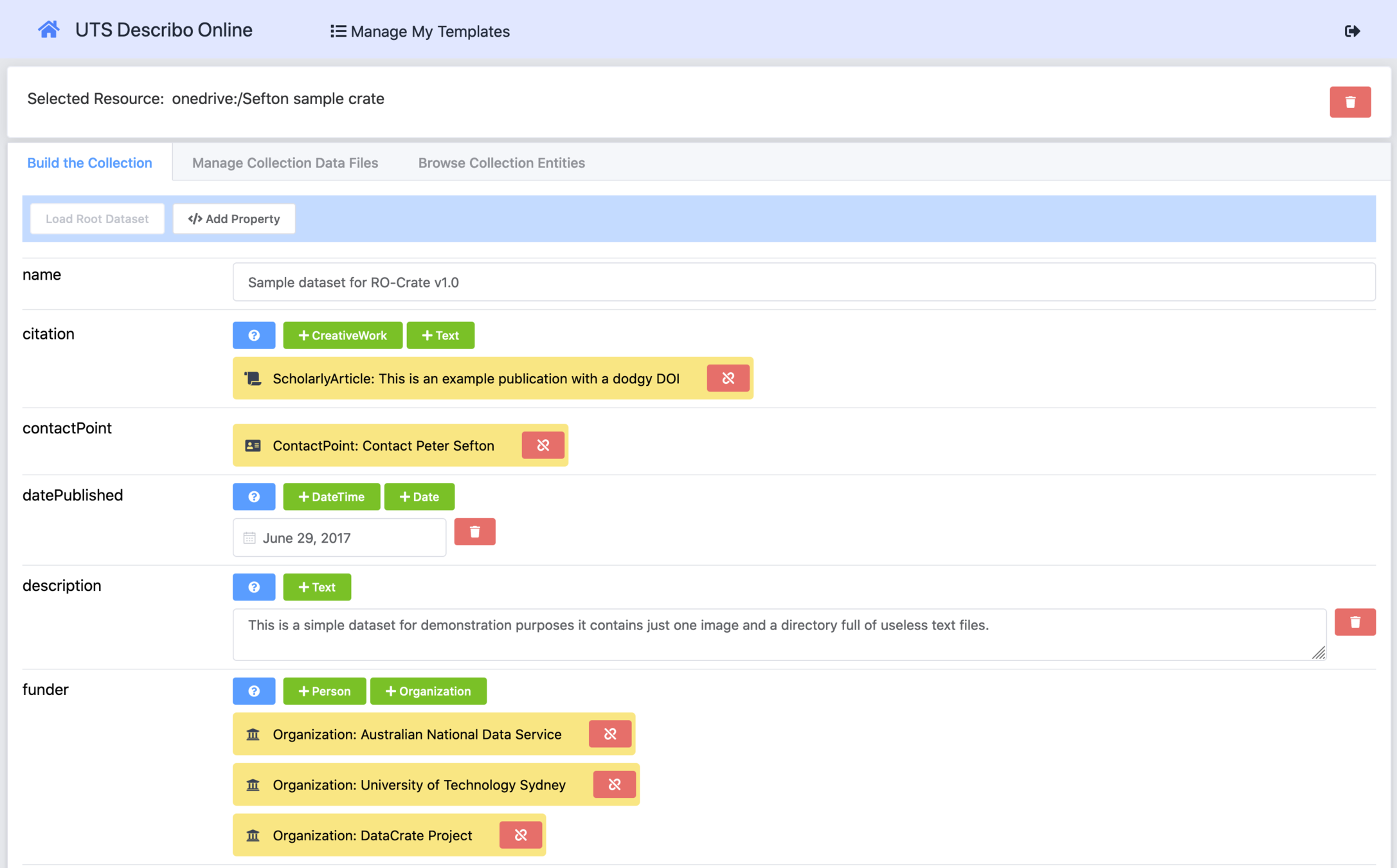
Making your own RO-Crate with Describo
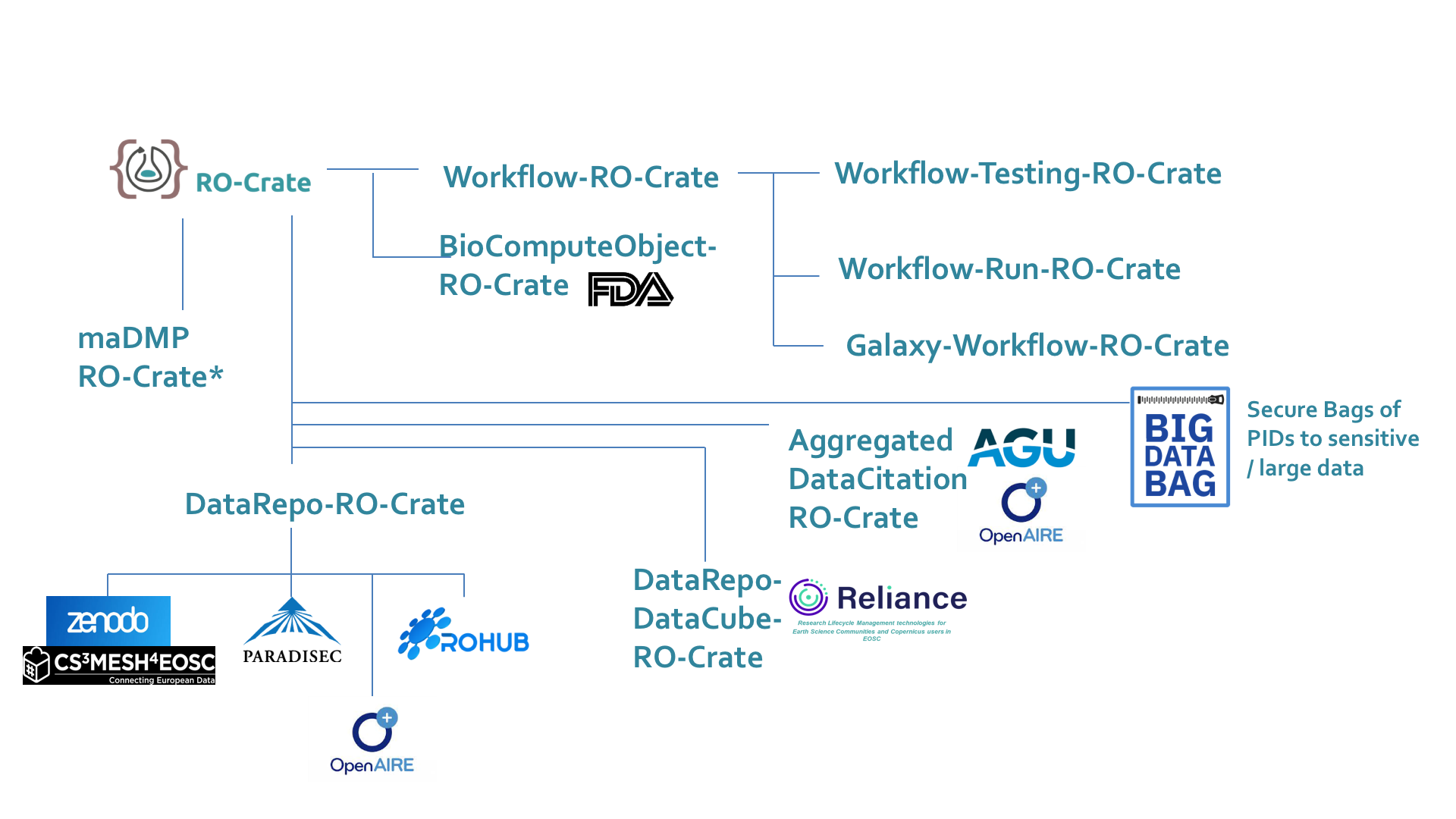
Multiple ways to use RO-Crate --> Profiles
Credit: Carole Goble
Dataverse Community Meeting 2021


Describing workflows
with RO-Crate











Containers
Describe workflow
Tests
Registry
Workflows
Authors and contributors
FAIR Digital Objects
…with RO-Crate as metadata object
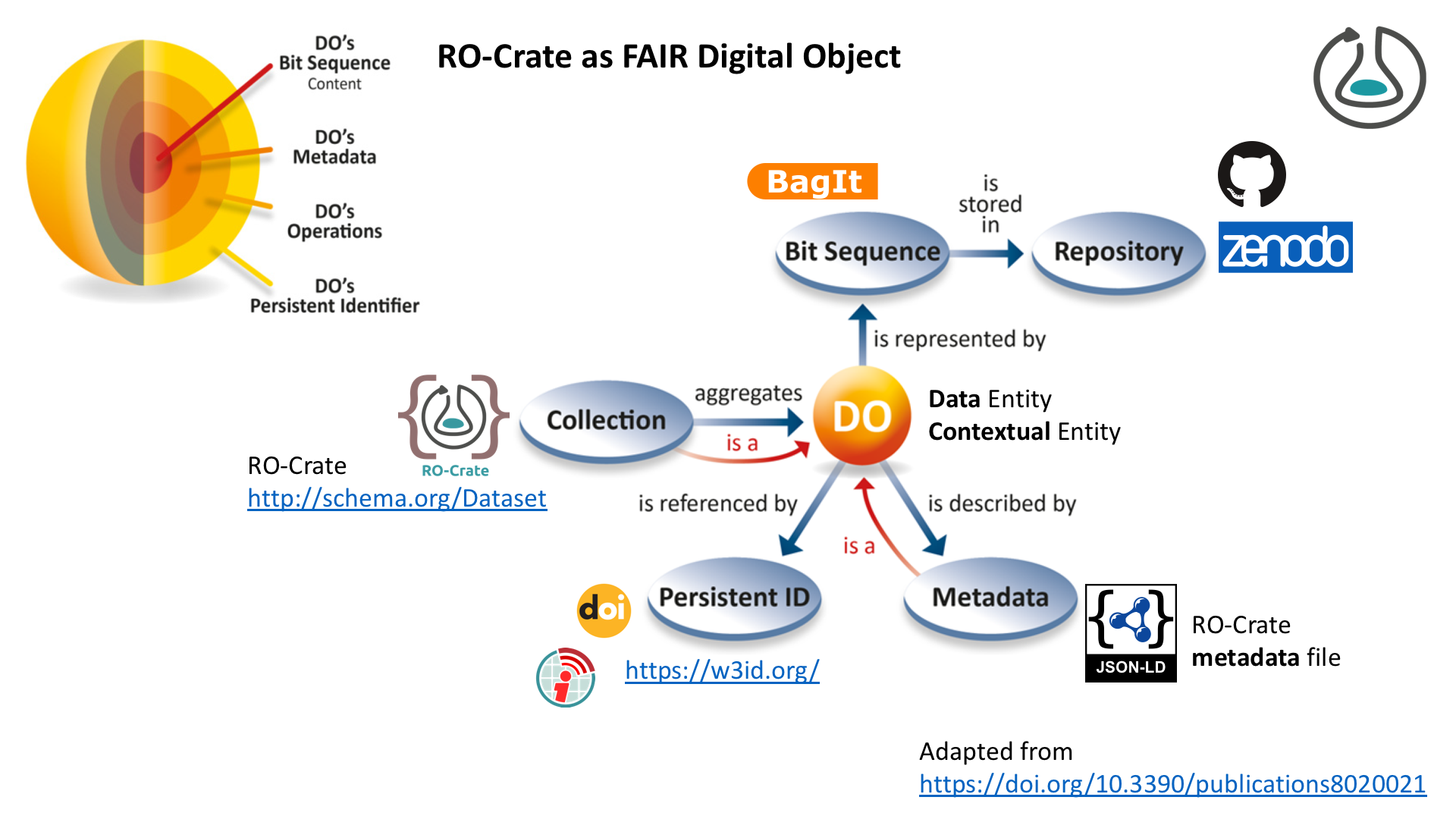
RO-Crate as FAIR Digital Object (FDO)
+ FAIR Signposting


Credit:
Herbert van de Sompel
FAIR Signposting: A KISS Approach to a Burning Issue
FAIR Signposting
Credit:
Herbert van de Sompel
FAIR Signposting: A KISS Approach to a Burning Issue
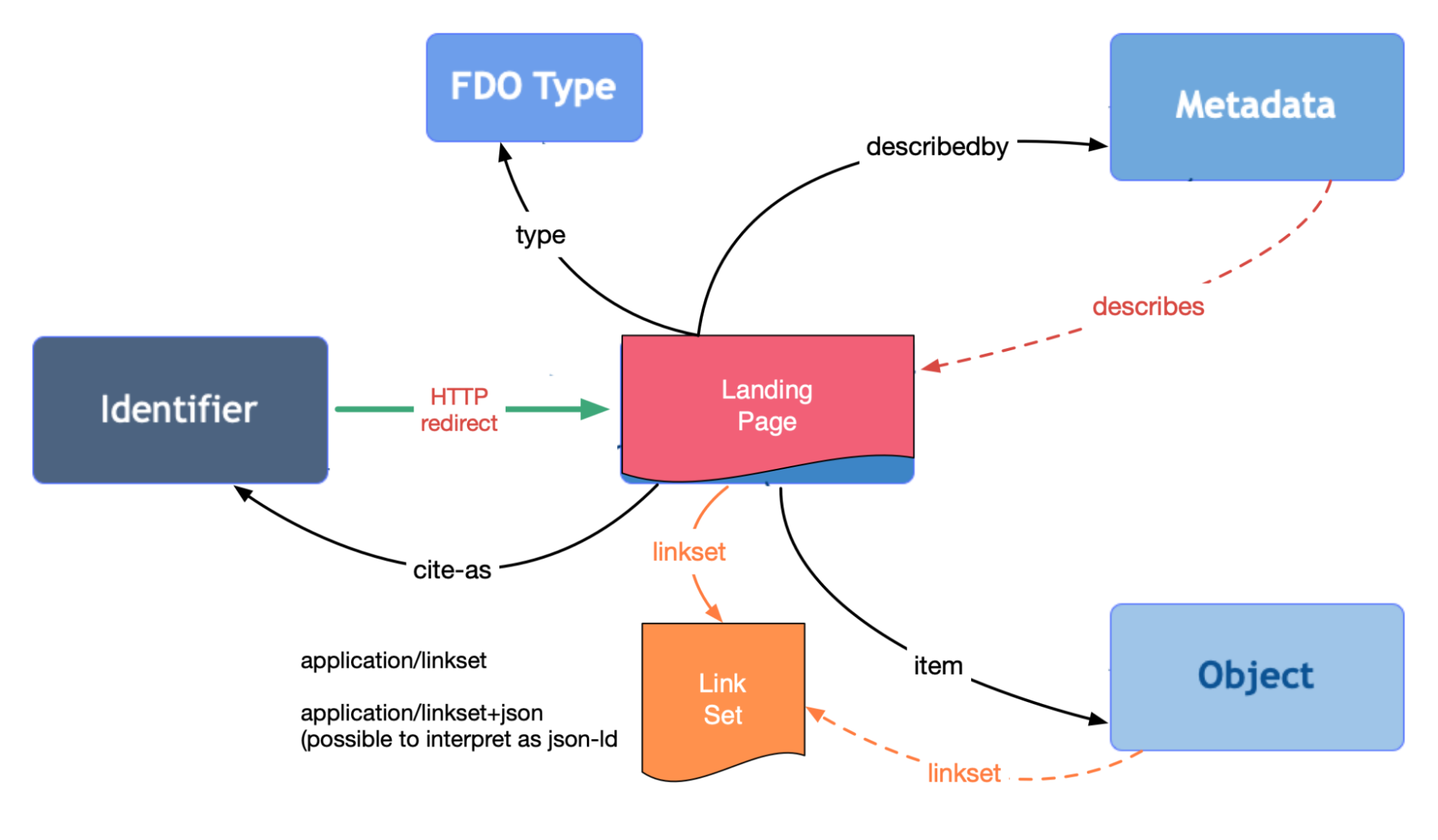
FAIR Signposting
HEAD https://workflowhub.eu/workflows/29?version=2 200 OK Link: <https://doi.org/10.48546/workflowhub.workflow.29.2>;rel=cite-as Link: <https://workflowhub.eu/workflows/29/ro_crate?version=2>;rel=describedby Link: <https://orcid.org/0000-0003-0513-0288>;rel=author
Link: <https://spdx.org/licenses/CC-BY-4.0>;rel=license
…
Next steps




workflowhub.eu and LifeMonitor
Workflow Run profile
Galaxy export of provenance
RO-Crate to submit to
COVID-19 Data Portal
Specimen Digital Refinery:
Digital twins as FDOs


Mass citation reliquary

RO-Crate for import/export
in general data repositories

Step-by-step
training material:
How to consume
many RO-Crates


Web interface to make
RO-Crates from cloud data

Data Cube for Earth Sciences
rohub.org

Canonical Workflow Building Blocks
CWLProv workflow provenance
--> RO-Crate

RO-Crate in Cloud API

Discussion
-
RO-Crate for workflows
-
RO-Crate in workflows
-
Linking Workflow-RO-Crates and data
-
Consuming data RO-Crates in workflows
-
-
Publishing and finding RO-Crates
-
FAIR Digital Objects (FDO) and FAIR Signposting
-
e.g. navigating from DOI to FAIR metadata
-
-
-
Training:
-
RO-Crates in ELIXIR RDM (RDMkit?)
-
RO-Crate FAIR Cookbook recipes?
-
Consumption of multiple RO-Crates
-
-
Existing communities and efforts
-
Using multiple EOSC metadata standards in RO-Crate
-
Formalizing RO-Crate profiles
(lightweight or using SHACL/ShEx/JSON Schema?)
-