FDO RO-Crate w/ Signposting
ELIXIR Biohackathon 2023 report
ELIXIR BioHackathon project #15
Claus Weiland
Jonas Grieb
Stian Soiland-Reyes
Leyla Jael Garcia
Sandy Rogers
Rohita Ravinder
Planning:
Herbert van de Sompel
Christophe Blanchi
"Enabling FAIR Digital Objects with RO-Crates, Signposting and Bioschemas"
# CHAPTER 2
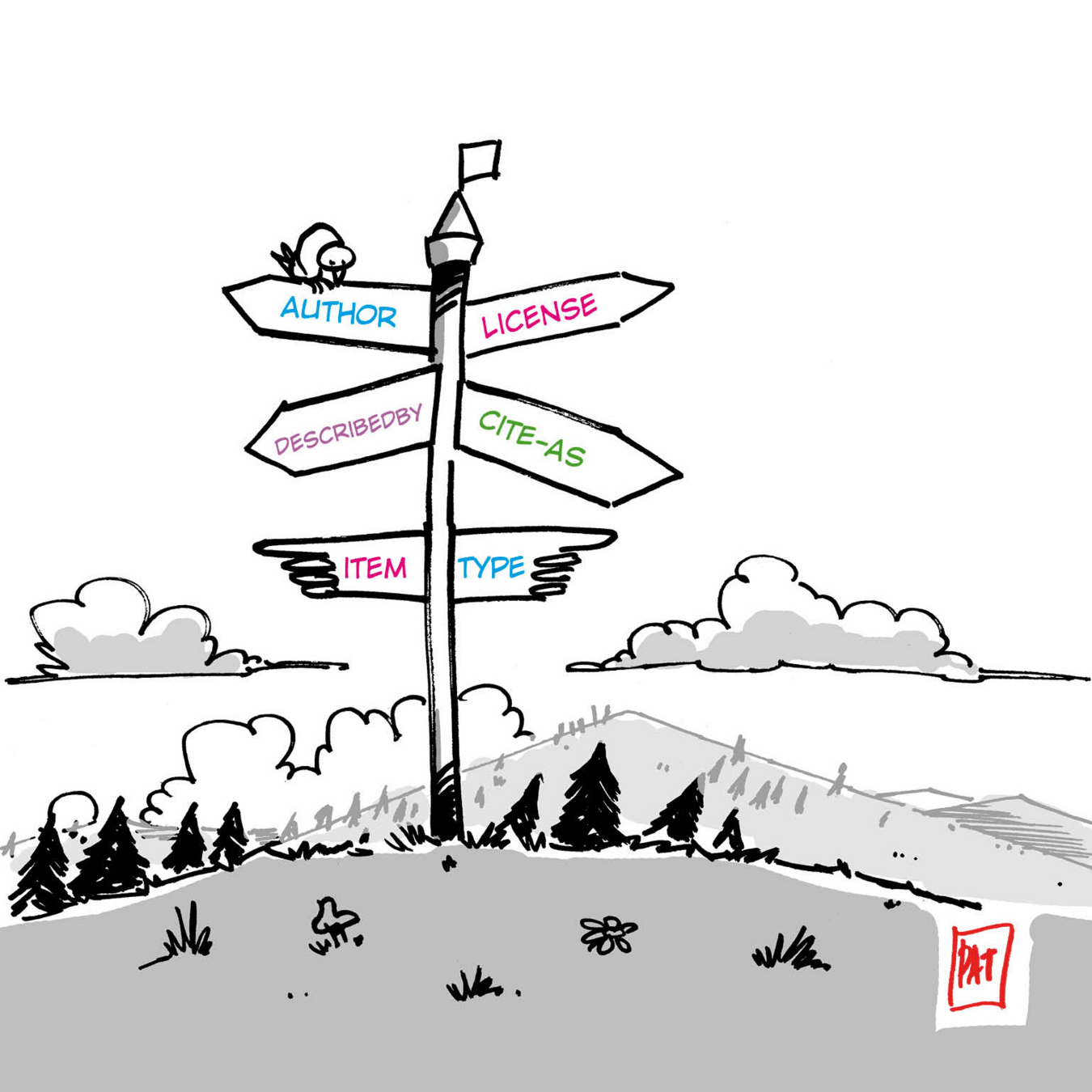
Goal #1: Signposting
Add Signposting Link headers to HTTP or HTML of existing repositories
Improve Signposting tooling
(base) stain@xena:~$ curl -I https://www.holofooddata.org/sample/SAMEA10104908 | grep Link
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
0 60095 0 0 0 0 0 0 --:--:-- 0:00:02 --:--:-- 0
Link: <https://www.holofooddata.org/api/samples/SAMEA10104908> ;
rel="describedby" ;
type="application/json" ;
profile="https://www.holofooddata.org/api/openapi.json",
<https://www.holofooddata.org/api/samples> ;
rel="collection" ;
type="application/json" ;
profile="https://www.holofooddata.org/api/openapi.json"
# PRESENTING CODE
Adding Signposting
Effectively miniature FDO
Profile links to OpenAPI specification

(base) stain@xena:~$ signposting https://www.holofooddata.org/sample/SAMEA10104908
Signposting for https://www.holofooddata.org/sample/SAMEA10104908
Collection: <https://www.holofooddata.org/api/samples>
DescribedBy: <https://www.holofooddata.org/api/samples/SAMEA10104908> application/json
# PRESENTING CODE
>>> import signposting
>>> signposting.find_signposting_http("https://www.holofooddata.org/sample/SAMEA10104908")
<Signposting context=https://www.holofooddata.org/sample/SAMEA10104908
collection=https://www.holofooddata.org/api/samples
describedBy=https://www.holofooddata.org/api/samples/SAMEA10104908>
>>> s = _
>>> s.
s.authors s.collection s.context_url s.for_context( s.license s.other_contexts s.types
s.citeAs s.context s.describedBy s.items s.linksets s.signposts
>>> s.describedBy
{<Signpost context=https://www.holofooddata.org/sample/SAMEA10104908 rel=describedby
target=https://www.holofooddata.org/api/samples/SAMEA10104908 type=application/json
profiles=https://www.holofooddata.org/api/openapi.json>}
>>> for d in s.describedBy:
... print(d.profiles)
...
frozenset({'https://www.holofooddata.org/api/openapi.json'})
Lessons learnt #1
Start with the easy
Signposting to your own custom JSON is better than no links at all.
Allow gradual growth to an FDO.
1.
2.
Identify the profile
Linking to an OpenAPI specification is much more powerful than just application/json.
Allow gradual building of an FDO profile.
3.
Annotate existing PIDs
Third-party services are free to express additional data about an official PID.
How to inform upstream about additional annotations?
# CHAPTER 2

# CHAPTER 2
Goal #2: RO-Crate
Simple deployment of RO-Crate on GitHub Pages

RO-Crate for a project
{
"@context": "https://w3id.org/ro/crate/1.1/context",
"@graph": [
{
"@id": "ro-crate-metadata.json",
"@type": "CreativeWork",
"conformsTo": {
"@id": "https://w3id.org/ro/crate/1.1"
},
"about": {
"@id": "./"
}
},
{
"@id": "./",
"@type": ["Dataset"],
"name": "RO-Crate for the research project 'Machine-actionable Software Management Plans'",
"description": "This RO-Crate describes the research project 'Machine-actionable Software Management Plans' including metadata about the project itself and the research outcomes created as part of it. The outcomes are listed via the hasPart property of the RO-crate. The purpose is providing a comprenhensive view of the research outcomes.",
"datePublished": "2023-11-02",
"license": {
"@type": "CreativeWork",
"@id": "https://spdx.org/licenses/CC-BY-4.0.html",
"name": "Creative Commons Attribution 4.0 International",
"alternateName": "CC BY 4.0",
"url": "https://creativecommons.org/licenses/by/4.0/"
},
"keywords": [
"machine-actionalibity", "software management plans", "rsearch software", "controlled vocabulary", "metadata schema", "ongoing", "SMP"
],
"about": { "@id": "https://zbmed-semtec.github.io/projects/2022_maSMP/"},
"hasPart": [
{"@id": "https://github.com/zbmed-semtec/maSMPs"},
{"@id": "https://doi.org/10.5281/zenodo.8089518"},
{"@id": "https://doi.org/10.52825/cordi.v1i.279"},
{"@id": "https://doi.org/10.5281/zenodo.8349183"},
{"@id": "https://doi.org/10.5281/zenodo.8087357"},
{"@id": "https://doi.org/10.4126/FRL01-006444988"},
{"@id": "https://ceur-ws.org/Vol-3415/paper-31.pdf"},
{"@id": "https://doi.org/10.4126/FRL01-006440396"},
{"@id": "https://doi.org/10.3897/rio.8.e94608"},
{"@id": "https://doi.org/10.5281/zenodo.7249674"}
]
},
{
"@id": "https://zbmed-semtec.github.io/projects/2022_maSMP",
"@type": "http://schema.org/ResearchProject",
"department": {
"@id": "https://zbmed-semtec.github.io/"
},
"description": "Our project corresponds to an extension of the RDA machine-actionable Data Management Plan (maDMP) application profile and its corresponding DMP Common Standard ontology (DCSO) in order to cover the case of ELIXIR Software Management Plans (SMP). Similar to DMPs, SMPs help formalize a set of structures and goals that ensure the software is accessible and reusable in the short, medium and long term. Although targeting the life sciences community, most of the elements of the ELIXIR SMPs are domain agnostic and could be used by other communities as well. DMPs and SMPs can be presented as text-based documents, sometimes guided by a set of questions corresponding to key points related to the lifecycle of either data or software. The RDA DMP Common Standards working group defined a maDMP to overcome limitations of text-based documents. We propose a similar path for the ELIXIR SMPs so they turn into machine-actionable SMPs (maSMPs).",
"foundingDate": {
"@type": "Date",
"@value": "2022-09-01"
},
"employee": [
{
"@id": "https://orcid.org/0000-0002-2910-7982"
},
{
"@id": "https://orcid.org/0000-0002-5037-0443"
},
{
"@id": "https://orcid.org/0009-0004-1529-0095"
},
{
"@id": "https://orcid.org/0000-0002-1018-0370"
},
{
"@id": "https://orcid.org/0000-0003-3986-0510"
}
],
"member": [
{
"@id": "https://orcid.org/0000-0003-0057-8788"
},
{
"@id": "https://orcid.org/0000-0001-8324-2897"
},
{
"@id": "https://orcid.org/0000-0002-3597-8557"
},
{
"@id": "https://orcid.org/0000-0002-2880-8947"
},
{
"@id": "https://orcid.org/0000-0002-0222-4273"
},
{
"@id": "https://orcid.org/0000-0002-0893-8509"
}
],
"alumni": [
{"@id": "https://orcid.org/0000-0003-2978-8922"}
],
"name": "Machine-Actionable Metadata for Software and Software Management Plans",
"funding": [
{
"@id": "https://ror.org/018mejw64"
},
{
"@id": "https://gepris.dfg.de/gepris/projekt/460234259"
},
{
"@id": "https://gepris.dfg.de/gepris/projekt/101017536"
}
],
"parentOrganization": [
{
"@id": "https://ror.org/0259fwx54"
}
],
"keywords": "ongoing, machine-actionability, software management plans, research software, controlled vocabulary, metadata schema",
"knowsAbout": [
{"@id": "https://github.com/zbmed-semtec/maSMPs"},
{"@id": "https://doi.org/10.5281/zenodo.8089518"},
{"@id": "https://doi.org/10.52825/cordi.v1i.279"},
{"@id": "https://doi.org/10.5281/zenodo.8349183"},
{"@id": "https://doi.org/10.5281/zenodo.8087357"},
{"@id": "https://doi.org/10.4126/FRL01-006444988"},
{"@id": "https://ceur-ws.org/Vol-3415/paper-31.pdf"},
{"@id": "https://doi.org/10.4126/FRL01-006440396"},
{"@id": "https://doi.org/10.3897/rio.8.e94608"},
{"@id": "https://doi.org/10.5281/zenodo.7249674"}
]
},
{
"@type": "SoftwareSourceCode",
"@id": "https://github.com/zbmed-semtec/maSMPs",
"datePublished": "2022-11-29",
"name": "maSMPs",
"description": "This project corresponds to an extension of the Research Data Alliance (RDA) machine-actionable Data Management Plan (maDMP) application profile and its corresponding DMP Common Standard ontology (DCSO) in order to cover the case of ELIXIR Software Management Plans (SMPs). Similar to DMPs, SMPs help formalize a set of structures and goals that ensure the software is accessible and reusable in the short, medium and long term. Although targeting the life sciences community, most of the elements of the ELIXIR SMPs are domain agnostic and could be used by other communities as well. DMPs and SMPs can be presented as text-based documents, sometimes guided by a set of questions corresponding to key points related to the lifecycle of either data or software. The RDA DMP Common Standards working group defined a maDMP to overcome limitations of text-based documents. We propose a similar path for the ELIXIR SMPs so they turn into machine-actionable SMPs (maSMPs).",
"url": "https://github.com/zbmed-semtec/maSMPs",
"citation": "Giraldo O, Geist L, Quiñones N, Solanki D, Rebholz-Schuhmann D, Castro LJ. machine-actionable Software Management Plan Ontology (maSMP Ontology). Zenodo; 2023. doi:10.5281/zenodo.8089518",
"programmingLanguage": [
"JavaScript", "Python"
]
},
{
"@type": "DefinedTermSet",
"@id": "https://doi.org/10.5281/zenodo.8089518",
"name": "machine-actionable Software Management Plan Ontology (maSMP Ontology)",
"description": "This is a metadata model in the form of an ontology representing the necessary metadata elements for a maSMP. The metadata model includes entities involved in software management planning; such as an SMP itself, software source code, software release, documentation, authors and their relations. We are reusing terms mainly from schema.org and from DCSO, with some few additions of our own.",
"url" : "https://zenodo.org/records/8089518",
"citation": "Giraldo Olga, Geist Lukas, Quiñones Nelson, Solanki Dhwani, Rebholz-Schuhmann Dietrich, & Castro Leyla Jael. (2023). machine-actionable Software Management Plan Ontology (maSMP Ontology) (1.0.0). Zenodo. https://doi.org/10.5281/zenodo.8089518",
"datePublished": "2023-06-28"
},
{
"@type": "DigitalDocument",
"@id": "https://doi.org/10.5281/zenodo.8349183",
"name": "Machine-Actionable Metadata for Software and Software Management Plans for NFDI - Presentation",
"@url": "https://zenodo.org/records/8349183",
"description": "Research data is on its way to be recognized as a first-class citizen in research; however, and despite its importance for science, software still has a long way to go. Recent initiatives are paving the way, including FAIR for Research Software and Software Management Plans. A step further towards machine-actionability is adding a structured metadata layer. Here we discuss some metadata elements useful to represent software and integrate it into management plans, and how it could be of benefit for NFDI.",
"citation": "Giraldo, O., Dessi, D., Dietze, S., Rebholz-Schuhmann, D., & Castro, L. J. (2023, September 15). Machine-Actionable Metadata for Software and Software Management Plans for NFDI - Presentation. Zenodo. https://doi.org/10.5281/zenodo.8349183",
"datePublished": "2022-09-15"
},
{
"@type": "Report",
"@id": "https://doi.org/10.5281/zenodo.8087357",
"name": "Workshop machine-actionable Software Management Plans",
"url": "https://zenodo.org/records/8087357",
"description": "The purpose of the workshop was validating the metadata schema support maSMPs [2, 3], improving its alignment with existing SMPs, and identifying gaps wrt ELIXIR SMPs and RDMO SMPs [4] and Research Data Alliance (RDA) maDMPs [5, 6]. The workshop counted with the participation of 10 people, from ELIXIR Software Development Best Practices Group, Bioschemas [7], RDMO SMPs, RDA DMP Common Standards Working Group, ARGOS and ZB MED. In addition to the resources represented by participants, we also include Codemeta [8] in the analysis. Here we present a report of what happened and what was achieved.",
"citation": "Giraldo, O., Cardoso, J., Martin del Pico, E., Gaignard, A., Geist, L., Grossmann, Y. V., Psomopoulos, F., Papadopoulou, E., Solanki, D., & Castro, L. J. (2023). Workshop machine-actionable Software Management Plans. Zenodo. https://doi.org/10.5281/zenodo.8087357",
"datePublished": "2023-06-27"
},
{
"@type": "ScholarlyArticle",
"@id": "https://doi.org/10.52825/cordi.v1i.279",
"conformsTo": "https://bioschemas.org/profiles/ScholarlyArticle/0.3-DRAFT",
"name": "Machine-Actionable Metadata for Software and Software Management Plans for NFDI",
"license": {
"@type": "CreativeWork",
"@id": "https://spdx.org/licenses/CC-BY-4.0.html",
"name": "Creative Commons Attribution 4.0 International",
"alternateName": "CC BY 4.0",
"url": "https://creativecommons.org/licenses/by/4.0/"
},
"datePublished": "2022-09-07"
},
{
"@type": "ScholarlyArticle",
"@id": "https://doi.org/10.4126/FRL01-006444988",
"conformsTo": "https://bioschemas.org/profiles/ScholarlyArticle/0.3-DRAFT",
"identifier": "DOI.10.4126/FRL01-006444988",
"name": "A metadata schema for machine-actionable Software Management Plans",
"datePublished": "2023-06-16"
},
{
"@type": "ScholarlyArticle",
"@id": "https://ceur-ws.org/Vol-3415/paper-12.pdf",
"conformsTo": "https://bioschemas.org/profiles/ScholarlyArticle/0.3-DRAFT",
"identifier": "CEUR:Vol-3415/paper-12",
"name": "A metadata analysis for machine-actionable Software Management Plans",
"datePublished": "2023-06-22"
},
{
"@type": "Poster",
"@id": "https://doi.org/10.4126/FRL01-006440396",
"name": "A metadata analysis for machine-actionable Software Management Plans - Poster",
"identifier": "DOI.10.4126/FRL01-006440396",
"datePublished": "2023-03-01"
},
{
"@type": "ScholarlyArticle",
"@id": "https://doi.org/10.3897/rio.8.e94608",
"conformsTo": "https://bioschemas.org/profiles/ScholarlyArticle/0.3-DRAFT",
"identifier": "DOI:10.3897/rio.8.e94608",
"name": "A FAIRification roadmap for ELIXIR Software Management Plans",
"datePublished": "2022-10-12"
},
{
"@type": "Poster",
"@id": "https://doi.org/10.5281/zenodo.7249674",
"name": "A FAIRification roadmap for ELIXIR Software Management Plans - Poster",
"identifier": "DOI.10.5281/zenodo.7249674",
"url": "https://doi.org/10.5281/zenodo.7249674",
"citation": "Olga Giraldo, Renato Alves, Dimitrios Bampalikis, Jose M Fernandez, Eva Martin del Pico, Fotis E Psomopoulos, Allegra Via, & Leyla Jael Castro. (2022). A FAIRification roadmap for ELIXIR Software Management Plans - Poster. First International Conference on FAIR Digital Objects (FDO2022). Zenodo. https://doi.org/10.5281/zenodo.7249674",
"datePublished": "2022-10-25"
},
{
"@id": "https://orcid.org/0000-0002-2910-7982",
"@type": "Person",
"givenName": "Lukas",
"familyName": "Geist"
},
{
"@id": "https://orcid.org/0000-0002-5037-0443",
"@type": "Person",
"givenName": "Nelson",
"familyName": "Quiñones"
},
{
"@id": "https://orcid.org/0009-0004-1529-0095",
"@type": "Person",
"givenName": "Dhwani",
"familyName": "Solanki"
},
{
"@id": "https://orcid.org/0000-0002-1018-0370",
"@type": "Person",
"givenName": "Dietrich",
"familyName": "Rebholz-Schuhmann"
},
{
"@id": "https://orcid.org/0000-0003-3986-0510",
"@type": "Person",
"givenName": "Leyla Jael",
"familyName": "Castro"
},
{
"@id": "https://orcid.org/0000-0003-0057-8788",
"@type": "Person",
"givenName": "Joao",
"familyName": "Cardoso"
},
{
"@id": "https://orcid.org/0000-0001-8324-2897",
"@type": "Person",
"givenName": "Eva",
"familyName": "Martin del Pico"
},
{
"@id": "https://orcid.org/0000-0002-3597-8557",
"@type": "Person",
"givenName": "Alban",
"familyName": "Gaignard"
},
{
"@id": "https://orcid.org/0000-0002-2880-8947",
"@type": "Person",
"givenName": "Yves Vincent",
"familyName": "Grossman"
},
{
"@id": "https://orcid.org/0000-0002-0222-4273",
"@type": "Person",
"givenName": "Fotis",
"familyName": "Psomopoulos"
},
{
"@id": "https://orcid.org/0000-0002-0893-8509",
"@type": "Person",
"givenName": "Elli",
"familyName": "Papadopoulou"
},
{
"@id": "https://orcid.org/0000-0003-2978-8922",
"@type": "Person",
"givenName": "Olga",
"familyName": "Giraldo"
},
{
"@id": "https://zbmed-semtec.github.io/",
"@type": "ResearchOrganization",
"name": "Semantic Technologies team at ZB MED"
},
{
"@id": "https://ror.org/018mejw64",
"@type": "Organization",
"alternateName": "German Research Foundation",
"name": "Deutsche Forschungsgemeinschaft",
"url": {
"@id": "http://www.dfg.de/en/"
}
},
{
"@id": "https://gepris.dfg.de/gepris/projekt/460234259",
"@type": "Grant",
"description": "Project no. 460234259 (corresponding to the NFDI4DataScience consortium)",
"funder": {
"@id": "https://ror.org/018mejw64"
},
"identifier": "460234259"
},
{
"@id": "https://gepris.dfg.de/gepris/projekt/460234259#project",
"@type": "Consortium",
"name": "NFDI4DataScience"
},
{
"@id": "https://gepris.dfg.de/gepris/projekt/101017536",
"@type": "Grant",
"description": "Grant agreement No 101017536, part of the Research Data Alliance and European Open Science Cloud Future call 2022",
"funder": {
"@id": "https://eoscfuture-grants.eu/"
},
"identifier": "101017536"
},
{
"@id": "https://ror.org/0259fwx54",
"@type": "ResearchOrganization",
"alternateName": "ZB MED Information Centre for Life Sciences",
"name": "Deutsche Zentralbibliothek für Medizin (ZB MED) - Informationszentrum Lebenswissenschaften",
"url": {
"@id": "https://zbmed.de/"
}
}
]
}# PRESENTING CODE

Lessons learnt #2
FDO RO-Crate scope
Decide on an crate size that fits available data/metadata and can have a resolvable identifier.
1.
2.
Digital twins are not datasets
For non-digital resources, the metadata needs separate PID so both can be described in the crate.
Use Detached Crate referencing only absolute URIs/PIDs.
3.
Prototyping
Allow FDO to be prototyped before the model is settled.
Simpler architectures (e.g. GitHub Pages) accelerates prototyping but have less flexibility.
# CHAPTER 2

# CHAPTER 2
Goal #3: FDO
Can we do hybrid deployment of FDOs with a Signposting/RO-Crate overlay?
Augment a portal that is built on Cordra

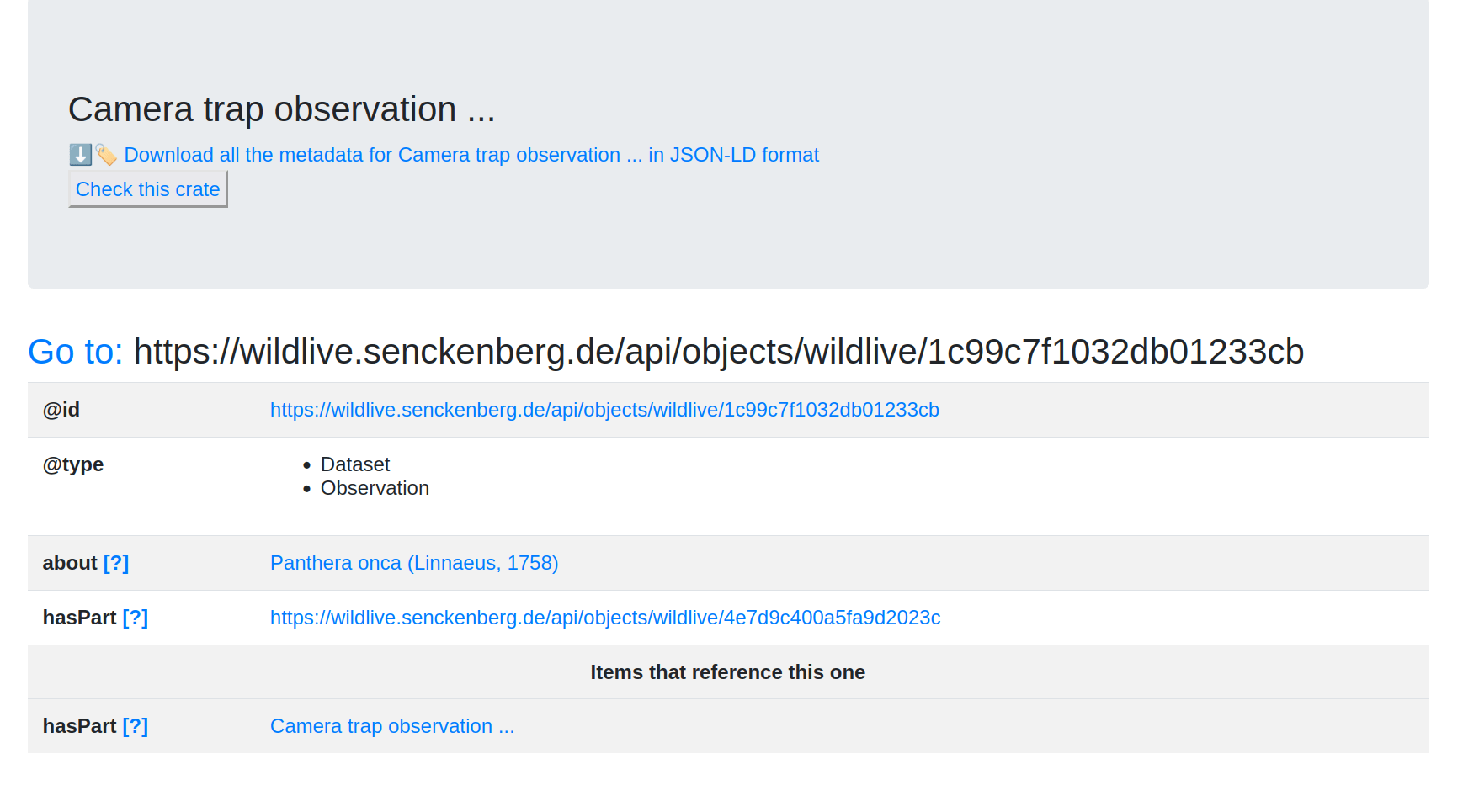
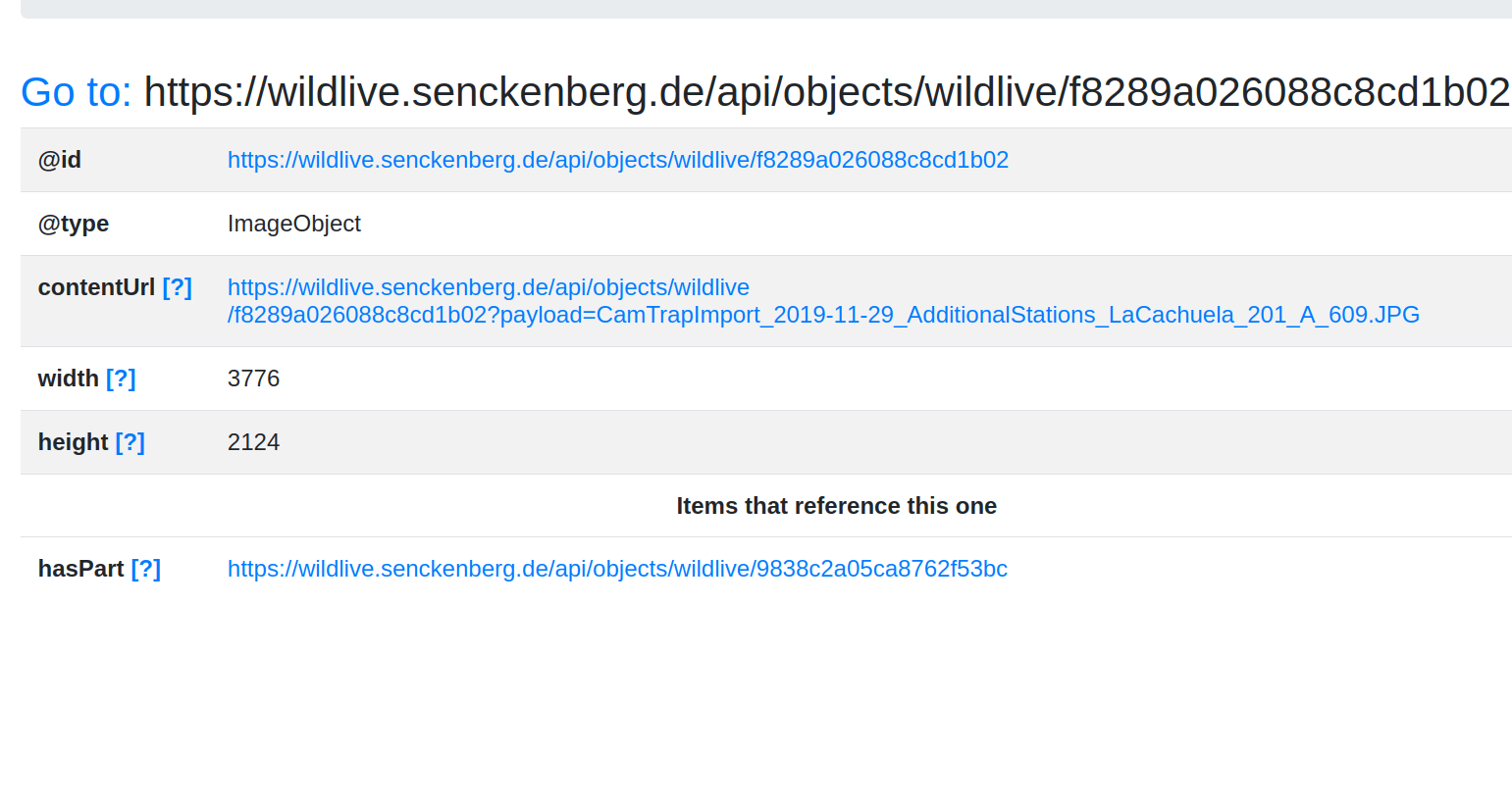

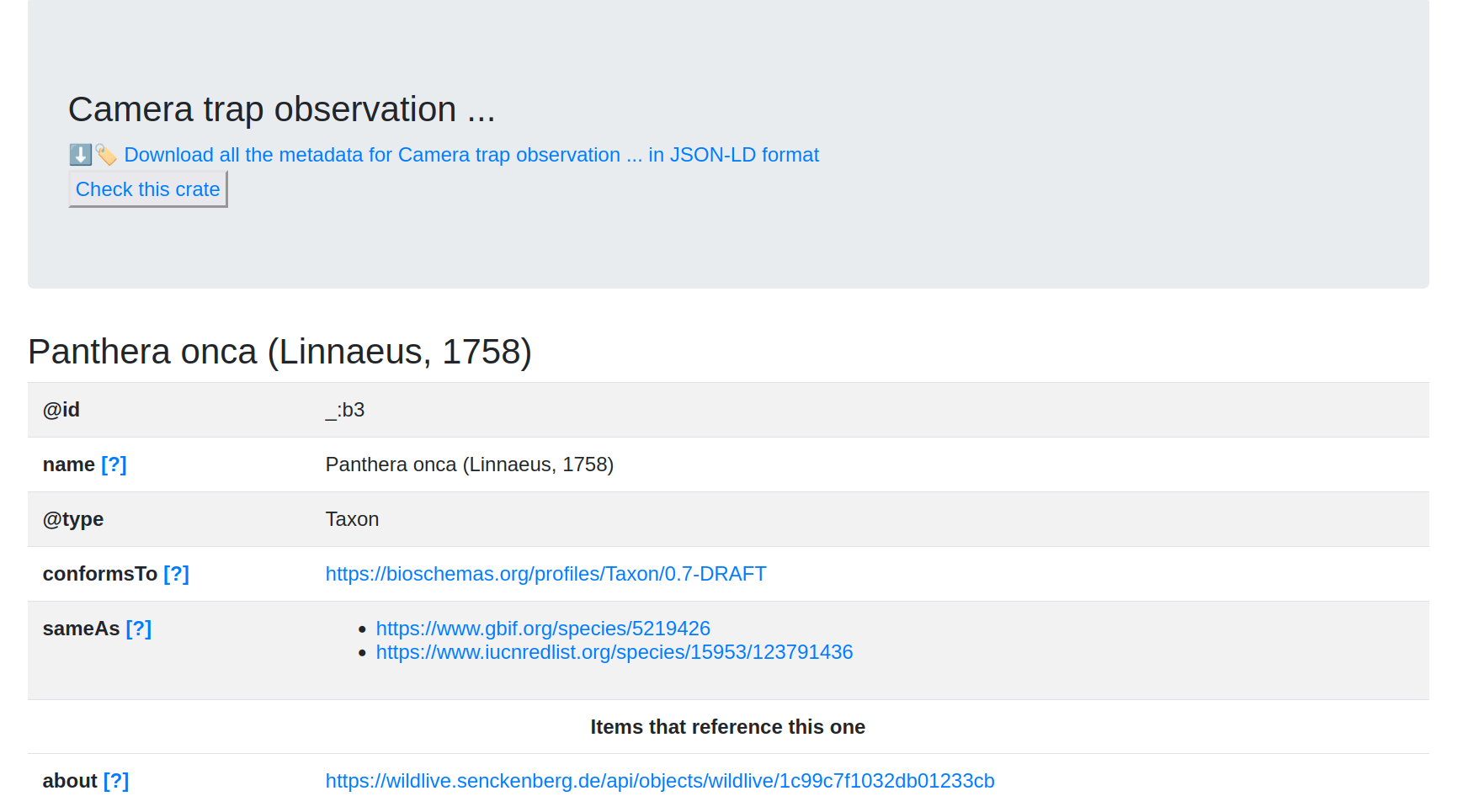
# PRESENTING CODE
Using Handle PIDs
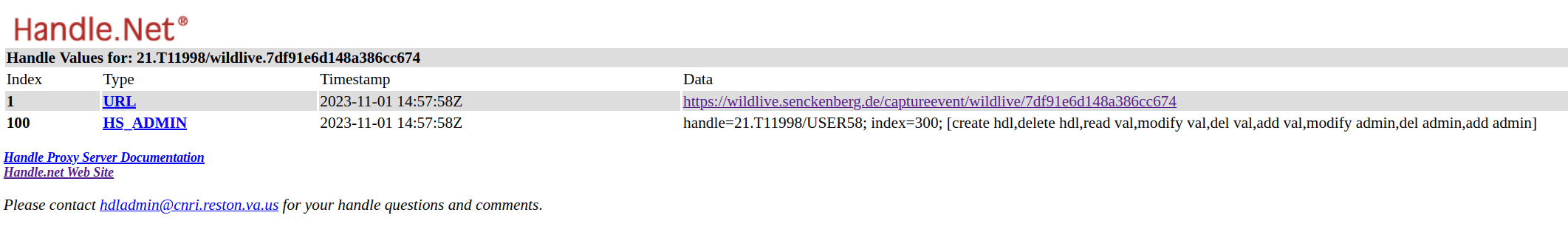

{
"@context": "https://w3id.org/ro/crate/1.1/context",
"@graph": [
{
"@id": "_:b0",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
{
"@id": "_:b1",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
{
"@id": "_:b2",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
{
"@id": "_:b3",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
{
"@id": "_:b4",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
{
"@id": "_:b5",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
{
"@id": "http://spdx.org/licenses/CC-BY-4.0"
},
{
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
{
"@id": "https://hdl.handle.net/21.T11998/wildlive.crate.7df91e6d148a386cc674",
"@type": "Dataset",
"author": {
"@id": "https://orcid.org/0000-0002-2631-4601",
"@type": "Person",
"name": "Martin Jansen"
},
"datePublished": "2023",
"hasPart": [
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/38a8bb080a5e48fdd309",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b0",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_606.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/9838c2a05ca8762f53bc",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b1",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_609.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/e0a97d39156c9b4b85c2",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b2",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_601.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/1c99c7f1032db01233cb",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b3",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_607.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/35d57b2098226936d146",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b4",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_605.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/4fbebc0422f047fa288e",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b5",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_608.JPG",
"height": 2124,
"width": 3776
}
}
],
"license": {
"@id": "http://spdx.org/licenses/CC-BY-4.0"
},
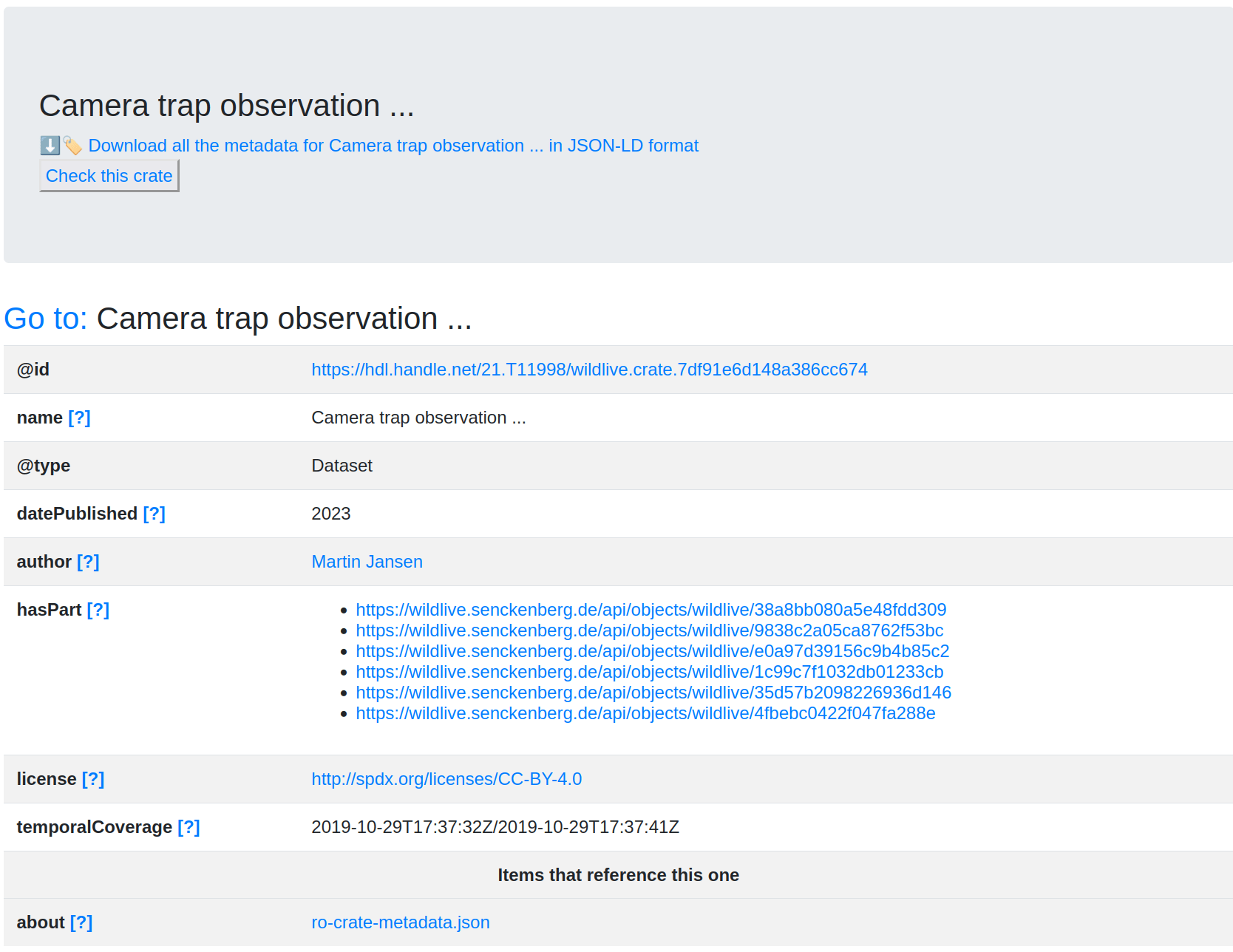
"name": "Camera trap observation ...",
"temporalCoverage": "2019-10-29T17:37:32Z/2019-10-29T17:37:41Z"
},
{
"@id": "https://orcid.org/0000-0002-2631-4601",
"@type": "Person",
"name": "Martin Jansen"
},
{
"@id": "https://w3id.org/ro/crate/1.1"
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/1c99c7f1032db01233cb",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b3",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_607.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/35d57b2098226936d146",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b4",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_605.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/38a8bb080a5e48fdd309",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b0",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_606.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_607.JPG",
"height": 2124,
"width": 3776
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/4fbebc0422f047fa288e",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b5",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_608.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_601.JPG",
"height": 2124,
"width": 3776
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/9838c2a05ca8762f53bc",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b1",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_609.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_608.JPG",
"height": 2124,
"width": 3776
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_605.JPG",
"height": 2124,
"width": 3776
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/e0a97d39156c9b4b85c2",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b2",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_601.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_609.JPG",
"height": 2124,
"width": 3776
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_606.JPG",
"height": 2124,
"width": 3776
},
{
"@id": "ro-crate-metadata.json",
"@type": "CreativeWork",
"conformsTo": {
"@id": "https://w3id.org/ro/crate/1.1"
},
"about": {
"@id": "https://hdl.handle.net/21.T11998/wildlive.crate.7df91e6d148a386cc674",
"@type": "Dataset",
"author": {
"@id": "https://orcid.org/0000-0002-2631-4601",
"@type": "Person",
"name": "Martin Jansen"
},
"datePublished": "2023",
"hasPart": [
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/38a8bb080a5e48fdd309",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b0",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/ffafa0893d4a2af6d0ba?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_606.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/9838c2a05ca8762f53bc",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b1",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/f8289a026088c8cd1b02?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_609.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/e0a97d39156c9b4b85c2",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b2",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/6f6afb2850b946bb9394?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_601.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/1c99c7f1032db01233cb",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b3",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/4e7d9c400a5fa9d2023c?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_607.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/35d57b2098226936d146",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b4",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/dbfaee3660bef31479a3?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_605.JPG",
"height": 2124,
"width": 3776
}
},
{
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/4fbebc0422f047fa288e",
"@type": [
"Dataset",
"Observation"
],
"about": {
"@id": "_:b5",
"@type": "Taxon",
"conformsTo": {
"@id": "https://bioschemas.org/profiles/Taxon/0.7-DRAFT"
},
"name": "Panthera onca (Linnaeus, 1758)",
"sameAs": [
"https://www.gbif.org/species/5219426",
"https://www.iucnredlist.org/species/15953/123791436"
]
},
"hasPart": {
"@id": "https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847",
"@type": "ImageObject",
"path": "https://wildlive.senckenberg.de/api/objects/wildlive/a98330d9df31a8d10847?payload=CamTrapImport_2019-11-29_AdditionalStations_LaCachuela_201_A_608.JPG",
"height": 2124,
"width": 3776
}
}
],
"license": {
"@id": "http://spdx.org/licenses/CC-BY-4.0"
},
"name": "Camera trap observation ...",
"temporalCoverage": "2019-10-29T17:37:32Z/2019-10-29T17:37:41Z"
}
}
]
}What are the profiles?
JSON Schema as FDO profile?
{
"identifier": "wildlive/e43b6bb9752e3f6aeeea",
"name": "CaptureEvent1.1.0",
"schema": {
"type": "object",
"title": "CaptureEvent",
"required": [
"dul:hasPart",
"time:hasBeginning",
"time:hasEnd"
],
"properties": {
"@id": {
"type": "string"
},
"@type": {
"type": "string"
},
"dul:hasPart": {
"type": "array",
"uniqueItems": true,
"items": {
"type": "string",
"title": "Observations",
"cordra": {
"type": {
"handleReference": {
"types": [
"Observation1.1.0"
]
}
}
}
}
},
"ods:capturedAt": {
"type": "string",
"cordra": {
"type": {
"handleReference": {
"types": [
"StationSetup1.1.0"
]
}
}
}
},
"time:hasBeginning": {
"type": "string",
"format": "date-time"
},
"time:hasEnd": {
"type": "string",
"format": "date-time"
},
"time:inXSDDateTimeStamp": {
"type": "string",
"format": "date-time"
}
}
},
"javascript": "exports.beforeSchemaValidation = beforeSchemaValidation;\nexports.onObjectResolution = onObjectResolution;\n\nconst CORDRA_BASE_URL = \"https://wildlive.senckenberg.de/api\";\nconst BASE_CONTEXT_URL =\n \"https://wildlive.senckenberg.de/api/objects/wildlive/basecontext.jsonld\";\n\nconst GBIF_TAXON_ID_MAPPING = {\n \"https://www.gbif.org/species/5219426\": [\n \"Jaguar\",\n \"Panthera onca (Linnaeus, 1758)\",\n \"https://www.iucnredlist.org/species/15953/123791436\",\n ],\n \"https://www.gbif.org/species/2441022\": [\n \"Aurochs\",\n \"Bos taurus Linnaeus, 1758\",\n ],\n \"https://www.gbif.org/species/2440995\": [\n \"Collared Peccary\",\n \"Pecari tajacu (Linnaeus, 1758)\",\n \"https://www.iucnredlist.org/species/41777/10562361\",\n ],\n};\n\nfunction contentToJsonLd(object) {\n object.content[\"@id\"] = object.id;\n const doType = object.type.replace(\"1.1.0\", \"\");\n let ldtype = \"ods:\" + doType;\n if (doType === \"Sensor\" || doType === \"Observation\") {\n ldtype = \"sosa:\" + doType;\n }\n object.content[\"@type\"] = ldtype;\n}\n\nfunction targetObjectToJsonLd(object) {\n contentToJsonLd(object);\n const ldcontext = object.content[\"@context\"];\n if (ldcontext !== undefined && Array.isArray(ldcontext)) {\n if (!ldcontext.includes(BASE_CONTEXT_URL)) {\n ldcontext.push(BASE_CONTEXT_URL);\n }\n } else {\n object.content[\"@context\"] = [BASE_CONTEXT_URL];\n }\n}\n\nfunction removeJsonLdFromContent(object) {\n if (\"@id\" in object.content) {\n delete object.content[\"@id\"];\n }\n if (\"@type\" in object.content) {\n delete object.content[\"@type\"];\n }\n // @context must be given ensured by beforeSchemaValidation\n const jsonldcontext = object.content[\"@context\"];\n /*if(jsonldcontext.length === 1){\n delete object.content[\"@context\"];\n } else {\n jsonldcontext.splice(jsonldcontext.indexOf(BASE_CONTEXT_URL),1);\n }*/\n}\n\nfunction beforeSchemaValidation(object, context) {\n if (!object.content) {\n throw \"Missing object content\";\n }\n const jsonldcontext = object.content[\"@context\"];\n if (!jsonldcontext) {\n throw \"Missing @context\";\n }\n if (!Array.isArray(jsonldcontext)) {\n throw \"Type error: @context must be of type array\";\n }\n if (!jsonldcontext.includes(BASE_CONTEXT_URL)) {\n throw (\n \"Error: The wildlive base context must be included in @context: \" +\n BASE_CONTEXT_URL\n );\n }\n removeJsonLdFromContent(object);\n}\n\nfunction onObjectResolutionBase(object) {\n targetObjectToJsonLd(object);\n return object;\n}\n\nfunction onObjectResolution(object, context) {\n onObjectResolutionBase(object);\n return object;\n}\nconst cordra = require(\"cordra\");\n\nexports.methods = {};\nexports.methods.getObjectAtGivenTime = getObjectAtGivenTime\nexports.methods.getObjectAtGivenTime.allowGet = true;\nexports.methods.getHierarchyInProject = getHierarchyInProject\nexports.methods.getHierarchyInProject.allowGet = true;\n\nexports.afterCreateOrUpdate = afterCreateOrUpdate;\nexports.beforeDelete = beforeDelete;\nexports.afterDelete = afterDelete;\n\nfunction afterCreateOrUpdate(object, context){\n afterCreateOrUpdateBase(object, context);\n}\n\nfunction afterCreateOrUpdateBase(object, context){\n const CordraLinker = Java.type(\"de.senckenberg.linkeddatacontroller.CordraLinker\");\n const cordraLinker = CordraLinker.instance();\n const objectStr = JSON.stringify(object);\n const contextStr = JSON.stringify(context);\n if (context.isNew) {\n cordraLinker.objectAfterCreate(objectStr, contextStr, \"1.1.0\");\n } else {\n const originalObjectStr = JSON.stringify(context.originalObject)\n cordraLinker.objectAfterUpdate(objectStr, originalObjectStr, contextStr, \"1.1.0\");\n }\n console.log(\"at the end of: afterCreateOrUpdate\");\n}\n\n\nfunction getObjectAtGivenTime(object, context) {\n const timestamp = context.params.timestamp;\n if(!timestamp){\n throw(\"Missing parameter: timestamp\");\n }\n const CordraLinker = Java.type(\"de.senckenberg.linkeddatacontroller.CordraLinker\");\n const cordraLinker = CordraLinker.instance();\n if(!object.content[\"ods:provObject\"]){\n throw(\"Object has no provenance\");\n }\n const version = cordraLinker.getObjectAtGivenTime(object.content[\"ods:provObject\"], timestamp);\n return JSON.parse(version);\n}\n\nfunction beforeDelete(object, context) {\n beforeDeleteBase(object, context);\n}\n\nfunction beforeDeleteBase(object, context) {\n const CordraLinker = Java.type(\"de.senckenberg.linkeddatacontroller.CordraLinker\");\n const cordraLinker = CordraLinker.instance();\n cordraLinker.deleteDependentLinkedObjectsBeforeDelete(JSON.stringify(object));\n}\n\nfunction afterDelete(object, context){\n // when userId is admin then it was probably called from internal deleting of\n // controlling linked digital object\n const CordraLinker = Java.type(\"de.senckenberg.linkeddatacontroller.CordraLinker\");\n const cordraLinker = CordraLinker.instance();\n if(context.userId !== \"admin\"){\n cordraLinker.afterDeleteReactiveBidirectionalLinking(JSON.stringify(object));\n }\n // NOTE: ProvenanceObjects should possibly not be deleted at all, but in the development\n // phase this is quite handy. Need to decide later whether to keep or remove this.\n const provenanceObjectId = object.content[\"ods:provObject\"];\n if(provenanceObjectId !== undefined)\n cordraLinker.afterDeleteProvenanceCleanup(provenanceObjectId);\n}\n\nfunction getHierarchyInProject(object, context){\n console.log(\"getHierarchyInProject \" + object.type)\n const CordraLinker = Java.type(\"de.senckenberg.linkeddatacontroller.CordraLinker\");\n const cordraLinker = CordraLinker.instance();\n const hierarchyString = cordraLinker.getHierarchyInProject(JSON.stringify(object));\n return JSON.parse(hierarchyString);\n}\n/* specific JavaScript functions for CaptureEvent */\nexports.methods.getDeserialized = getDeserialized;\nexports.methods.getDeserializedContent = getDeserializedContent;\nexports.methods.getDeserialized.allowGet = true;\nexports.methods.getDeserializedContent.allowGet = true;\nexports.methods.getDeserializedImages = getDeserializedImages;\nexports.methods.getDeserializedImages.allowGet = true;\nexports.methods.getAsROCrate = getAsROCrate;\nexports.methods.getAsROCrate.allowGet = true;\nexports.onObjectResolution = onObjectResolution;\n\nfunction getDeserialized(object, context) {\n return deserializeCaptureEvent(object, false);\n}\n\nfunction getDeserializedContent(object, context) {\n return deserializeCaptureEvent(object, true);\n}\n\nfunction getDeserializedImages(object, context) {\n return deserializeCaptureEvent(object, false, false);\n}\n\nfunction deserializeCaptureEvent(\n object,\n contentOnly,\n includeAnnotations = true\n) {\n let deserialized;\n const author = {\n \"@id\": \"https://orcid.org/0000-0002-2631-4601\",\n \"@type\": \"schema:Person\",\n \"schema:name\": \"Martin Jansen\",\n };\n if (contentOnly) {\n deserialized = JSON.parse(JSON.stringify(object.content));\n deserialized[\"schema:license\"] = \"http://spdx.org/licenses/CC-BY-4.0\";\n deserialized[\"schema:author\"] = author;\n } else {\n deserialized = JSON.parse(JSON.stringify(object));\n deserialized.content[\"schema:license\"] =\n \"http://spdx.org/licenses/CC-BY-4.0\";\n deserialized.content[\"schema:author\"] = author;\n }\n const observationIds = object.content[\"dul:hasPart\"];\n if (Array.isArray(observationIds)) {\n const observations = [];\n for (let i = 0; i < observationIds.length; i++) {\n const observation = cordra.get(observationIds[i]);\n const mediaId = observation.content[\"sosa:hasResult\"];\n if (mediaId !== undefined) {\n const mediaObject = cordra.get(mediaId);\n const annotations = [];\n const annotationIDs = mediaObject.content[\"ods:hasAnnotations\"];\n if (Array.isArray(annotationIDs) && includeAnnotations) {\n for (let j = 0; j < annotationIDs.length; j++) {\n const annotation = cordra.get(annotationIDs[j]);\n\n const annotationCommmentsIDs =\n annotation.content[\"ods:hasAnnotations\"];\n if (Array.isArray(annotationCommmentsIDs)) {\n annotation.content[\"comments\"] = [];\n\n for (\n let annotationCommmentsIDsIDX = 0;\n annotationCommmentsIDsIDX < annotationCommmentsIDs.length;\n annotationCommmentsIDsIDX++\n ) {\n annotation.content.comments.push({\n id: annotationCommmentsIDs[annotationCommmentsIDsIDX],\n comment: cordra.get(\n annotationCommmentsIDs[annotationCommmentsIDsIDX]\n ).content[\"oa:hasBody\"].comment,\n });\n }\n }\n\n if (contentOnly) {\n annotations.push(annotation.content);\n } else {\n annotations.push(annotation);\n }\n }\n mediaObject.content[\"ods:hasAnnotations\"] = annotations;\n }\n if (contentOnly) {\n observation.content[\"sosa:hasResult\"] = mediaObject.content;\n } else {\n observation.content[\"sosa:hasResult\"] = mediaObject;\n }\n }\n if (contentOnly) {\n observations.push(observation.content);\n } else {\n observations.push(observation);\n }\n }\n if (contentOnly) {\n deserialized[\"dul:hasPart\"] = observations;\n } else {\n deserialized.content[\"dul:hasPart\"] = observations;\n }\n }\n return deserialized;\n}\n\n/* overrides default beforeDelete behavior */\nfunction beforeDelete(object, context) {\n if (context.userId !== \"admin\") {\n const observations = object.content[\"dul:hasPart\"];\n if (Array.isArray(observations) && observations.length > 1) {\n throw \"Error: A CaptureEvent can only be deleted when it has no observations in /dul:hasPart\";\n }\n }\n}\n\nfunction getAsROCrate(object, context) {\n let landingPageUrl =\n \"https://wildlive.senckenberg.de/captureevent/\" + object.id;\n if (object.id === \"wildlive/7df91e6d148a386cc674\") {\n landingPageUrl =\n \"https://hdl.handle.net/21.T11998/wildlive.crate.7df91e6d148a386cc674\";\n }\n const baseUrl = \"https://wildlive.senckenberg.de/api/objects/\";\n const rocrate = {\n \"@context\": \"https://w3id.org/ro/crate/1.1/context\",\n \"@graph\": [\n {\n \"@type\": \"CreativeWork\",\n \"@id\": \"ro-crate-metadata.json\",\n conformsTo: { \"@id\": \"https://w3id.org/ro/crate/1.1\" },\n about: { \"@id\": landingPageUrl },\n },\n {\n \"@id\": landingPageUrl,\n \"@type\": \"Dataset\",\n datePublished: \"2023\",\n hasPart: [],\n name: \"Camera trap observation ...\",\n license: { \"@id\": \"http://spdx.org/licenses/CC-BY-4.0\" },\n author: {\n \"@id\": \"https://orcid.org/0000-0002-2631-4601\",\n },\n },\n {\n \"@id\": \"https://orcid.org/0000-0002-2631-4601\",\n \"@type\": \"Person\",\n name: \"Martin Jansen\",\n },\n ],\n };\n\n const cecontent = object.content;\n if (\n cecontent[\"time:hasBeginning\"] !== undefined &&\n cecontent[\"time:hasEnd\"] !== undefined\n ) {\n const iSO8601TimeInterval =\n cecontent[\"time:hasBeginning\"] + \"/\" + cecontent[\"time:hasEnd\"];\n rocrate[\"@graph\"][1][\"temporalCoverage\"] = iSO8601TimeInterval;\n }\n const observationIds = object.content[\"dul:hasPart\"];\n if (Array.isArray(observationIds)) {\n const observations = [];\n for (let i = 0; i < observationIds.length; i++) {\n const observation = cordra.get(observationIds[i]);\n const mediaId = observation.content[\"sosa:hasResult\"];\n const observationUrl = baseUrl + observation.id;\n if (mediaId !== undefined) {\n const mediaObject = cordra.get(mediaId);\n const imcontent = mediaObject.content;\n const imageUrl = baseUrl + mediaObject.id;\n const roObservation = {\n \"@id\": observationUrl,\n \"@type\": [\"Dataset\", \"Observation\"],\n hasPart: { \"@id\": imageUrl },\n };\n\n const annotationIDs = mediaObject.content[\"ods:hasAnnotations\"];\n if (Array.isArray(annotationIDs)) {\n for (let j = 0; j < annotationIDs.length; j++) {\n const annotation = cordra.get(annotationIDs[j]);\n const body = annotation.content[\"oa:hasBody\"];\n if (\n typeof body === \"object\" &&\n !Array.isArray(body) &&\n body !== null &&\n body[\"@type\"] === \"ods:TaxonomicBody\" &&\n typeof body[\"dwc:taxonID\"] === \"string\" &&\n body[\"dwc:taxonID\"].length > 0\n ) {\n taxon_mappings = GBIF_TAXON_ID_MAPPING[body[\"dwc:taxonID\"]];\n const taxon = {\n \"@type\": \"Taxon\",\n name: taxon_mappings[1],\n sameAs: [body[\"dwc:taxonID\"]],\n taxonRank: [\n \"http://rs.tdwg.org/ontology/voc/TaxonRank#Species\",\n \"http://www.wikidata.org/entity/Q7432\",\n \"species\",\n ],\n conformsTo: {\n \"@id\": \"https://bioschemas.org/profiles/Taxon/0.7-DRAFT\",\n },\n };\n if (taxon_mappings.length === 3) {\n taxon.sameAs.push(taxon_mappings[2]);\n }\n if (!Array.isArray(roObservation.about)) {\n roObservation.about = [taxon];\n } else {\n roObservation.about.push(taxon);\n }\n }\n }\n }\n\n rocrate[\"@graph\"].push(roObservation);\n rocrate[\"@graph\"][1][\"hasPart\"].push({ \"@id\": observationUrl });\n const roImageObject = {\n \"@id\": imageUrl,\n \"@type\": \"ImageObject\",\n contentUrl: imcontent[\"ods:mediaURL\"],\n };\n if (imcontent[\"ods:mediaWidth\"] !== undefined) {\n roImageObject[\"width\"] = imcontent[\"ods:mediaWidth\"];\n }\n if (imcontent[\"ods:mediaHeight\"] !== undefined) {\n roImageObject[\"height\"] = imcontent[\"ods:mediaHeight\"];\n }\n rocrate[\"@graph\"].push(roImageObject);\n }\n }\n }\n\n return rocrate;\n}\n\n/* overrides onObjectResolution from base */\nfunction onObjectResolution(object, context) {\n onObjectResolutionBase(object);\n object.content[\"schema:license\"] = \"http://spdx.org/licenses/CC-BY-4.0\";\n object.content[\"schema:author\"] = {\n \"@id\": \"https://orcid.org/0000-0002-2631-4601\",\n \"@type\": \"schema:Person\",\n \"schema:name\": \"Martin Jansen\",\n };\n return object;\n}\n",
"authConfig": {
"defaultAclRead": [
"public"
],
"defaultAclWrite": [
"creator"
],
"aclCreate": [
"wildlive/imageprovider"
],
"aclMethods": {
"instance": {
"getHierarchyInProject": [
"readers"
],
"getDeserialized": [
"readers"
],
"getDeserializedContent": [
"readers"
],
"getDeserializedImages": [
"readers"
],
"getAsROCrate": [
"readers"
]
}
}
}
}BioDT kernel attributes
Kernel Attributes Table
IndexPropertyDescriptionTypeCardinalityCommentsExample
| 1 | profile | The FDO profile that a certain metadata description follows. | URL | 1/1 | For now, we can refer to this repo for the profiles. | |
| 2 | license | License for the FDO the metadata description is about. | URL | 1/1 | The FDO might have a certain license, however, the metadata file should have a CC0 license. | "http://creativecommons.org/licenses/by/4.0/" |
| 3 | type | The nature of the object the metadata describes. For now, this follows the RO-Crate conventions. | Text or URL | 1/many | "Dataset" | |
| 4 | identifier | An identifier for the FDO. Ideally, a globally unique, persistent and resolvable identifier. | Text or URL | 1/1 | "https://doi.org/10.1111/1365-2664.12222" | |
| 5 | name | Name of the FDO. | Text | 1/many | "BEEHAVE model" | |
| 6 | description | Short description of what the FDO is. | Text | 1/many | "BEEHAVE is a computer model to simulate the development of a honeybee colony and its nectar and pollen foraging behavior in different..." | |
| 7 | datePublished | Date of publication of the FDO. | Date | 1/1 | "2014-03-04" | |
| 8 | dateModified | Most recent date where the FDO was modified. | Date | 1/1 | "2014-03-04" | |
| 9 | author | Author of the FDO. | Person or Organization | 1/many | If possible, use an ORCiD, otherwise use full name in natural order. | "https://orcid.org/0000-0003-0791-7164" |
DISSCo kernel attributes
|
Index |
attribute |
example |
Cardinality |
type |
|
1 |
fdoProfile PID to a machine readable description of the attributes in the FDO record |
1/1 |
pid |
|
|
2 |
fdoRecordLicense the licence for the FDO record, required to be always public domain |
1/1 |
string |
|
|
3 |
digitalObjectType PID to a description of the Type of digital object that defines the metadata, bit sequences (if any) and operations for the object |
https://hdl.handle.net/21.T11148/894b1e6cad57e921764e |
1/1 |
pid |
|
4 |
digitalObjectName name of the object type for humans |
digital specimen type 1 |
1/1 |
string |
|
5 |
pid the PID of which the FDO record is part, in DiSSCo this is a Handle or DOI. It is recommended to store this pid also in the local collection management system for the specimen. |
https://doi.org/10.22/GEE-W3J-HL2 |
1/1 |
pid |
|
6 |
pidIssuer In case of a DOI this is a PID for the DOI Registration Agency |
https://hdl.handle.net/10.17183 |
1/1 |
pid |
|
7 |
pidIssuerName |
DataCite |
1/1 |
string |
|
8 |
issuedForAgent In the case of a digital specimen, this is a PID for DiSSCo as the agent responsible for serving the digital specimen object |
1/1 |
pid |
|
|
9 |
issuedForAgentName |
DiSSCo |
1/1 |
string |
|
10 |
pidRecordIssueDate date the pid record was created |
2022-11-24 |
1/1 |
date |
|
11 |
pidRecordIssueNumber starts with 1 and is incrementally increased by 1 every time the pid record is updated. Compatible with DOI schema requirements. |
2 |
1/1 |
int |
|
12 |
structuralType Nature of the digital object, compatible with DOI schema requirements. The nature of a digital specimen object is always "digital". Other digital objects (outside DiSSCo) could be of physical, performance or abstraction nature. |
digital |
1/1 |
literal |
|
13 |
pidStatus A PID is considered to have a lifecycle, PID status indicates the status in the life cycle, e.g. draft, active, retired. PID statuses are described further in the PID infrastructure design. One of: one of: DRAFT, ACTIVE, RETIRED, OBSOLETE, FAILED, MERGED, SPLIT |
DRAFT |
1/1 |
vocab |
|
|
||||
|
40 |
referentType A generic name for the type of object that the DOI refers to. This is different from digitalObjectType that points to a specific type, e.g. there can be different types of digital specimens that each have a slightly different metadata schema because they describe a different kind of specimen, like a botanical versus a geological specimen. |
digital specimen |
1/1 |
literal |
|
41 |
referentDoiName the bare DOI Name string for the PID, e.g. without the resolver. |
10.22/GEE-W3J-HL2 |
1/1 |
string |
|
42 |
referentName In the case of a digital specimen this is the name for the object in the collection, which can be anything from a taxon name to a collection number. |
Mus musculus type 1 |
1/1 |
string |
|
43 |
primaryReferentType The primary type of the referent in the DOI Kernel XML Schema (e.g. creation, party, event). This is an open list. For digital specimens and media it will always be creation. |
creation |
1/1 |
literal |
- Examples are using test prefixes
- Profile handles don't have a profile nor type
- Index seems to have meaning?
- Mixes general and specific
- +1 Use of full URIs
- +1 type AND profile
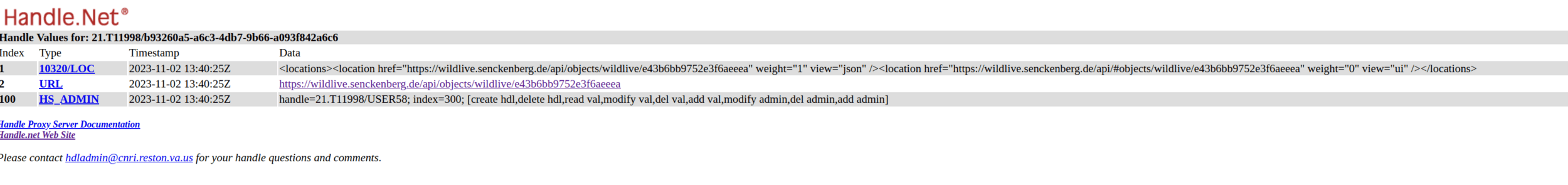
Handling handle
- Requesting EOSC B2Handle service is very slow
- Was not assigned "real" prefixes -- common issue
- B2Handle (test) server unreliable:
- SSL errors
- Could not update PIDs
- Unclear how to indicate profile in handle
- Unclear how much of the FDO metadata should be duplicated into the Handle
- Unclear which keys to use in Handle
- Unclear if index in Handle matters (FDO profile is index 1?)
Lessons learnt #3
User in focus
Make sure human browsers get HTML (use 10320/LOC)
Use full https://hdl.handle.net/ URIs
1.
2.
Profile profile needed
Still too unclear how to express a profile. JSON Schema kind-a works but is rudimentary.
RO-Crate specify (sub)profile inside -- do we duplicate that in its Handle?
3.
Granularity varies
The FDOs may be fine-grained (e.g. to support their operations)
An RO-Crate should make sense on its own -- augment with metadata from compound FDOs.
# CHAPTER 2



