Stochastic Morphometry and
Center for Computational Evolutionary Morphometrics
Stefan Sommer, Rasmus Nielsen, Christy Hipsley, Mads Nielsen


UCPH Data+

Phylogenetics

Darwin 1859
Phylogenetics - Estimating Trees
Until ca. 1980: morphological characters
After ca. 1980: DNA
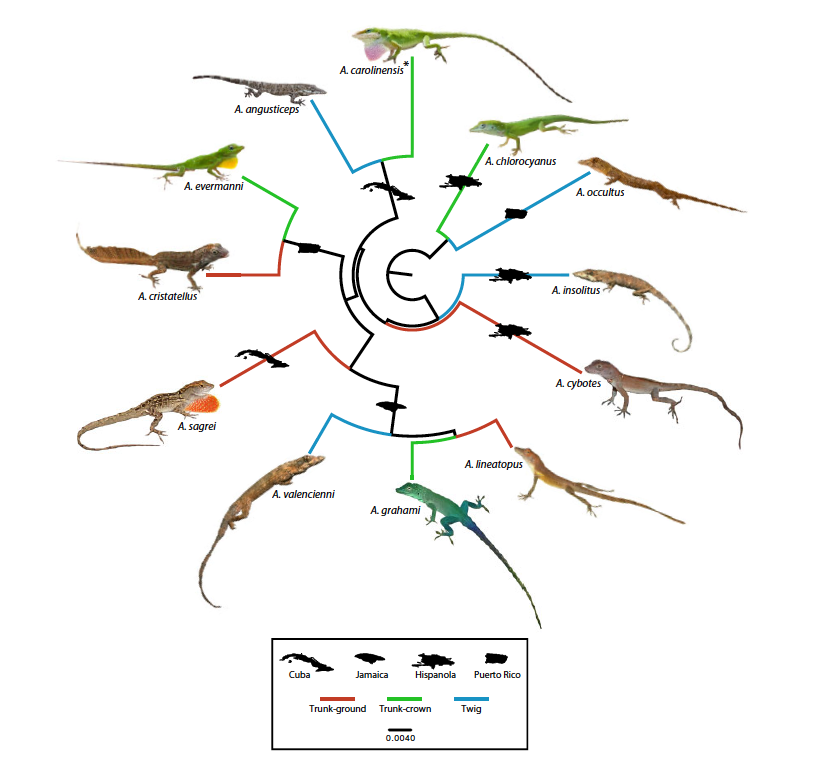
Corbett-Deitig et al. MBE 37: 160-1614
Estimating trees using DNA
- Modeling evolution using Markov chain with state space = {A, C, T, G}.
- Transition probabilities of the process is calculated by exponentiating infinitesimal rate matrix.
- Likelihood calculated using Felsenstein’s pruning algorithm and then numerically optimized or used in Bayesian inference.
- Genomic sequencing provides millions of observations for accurate estimation of trees.
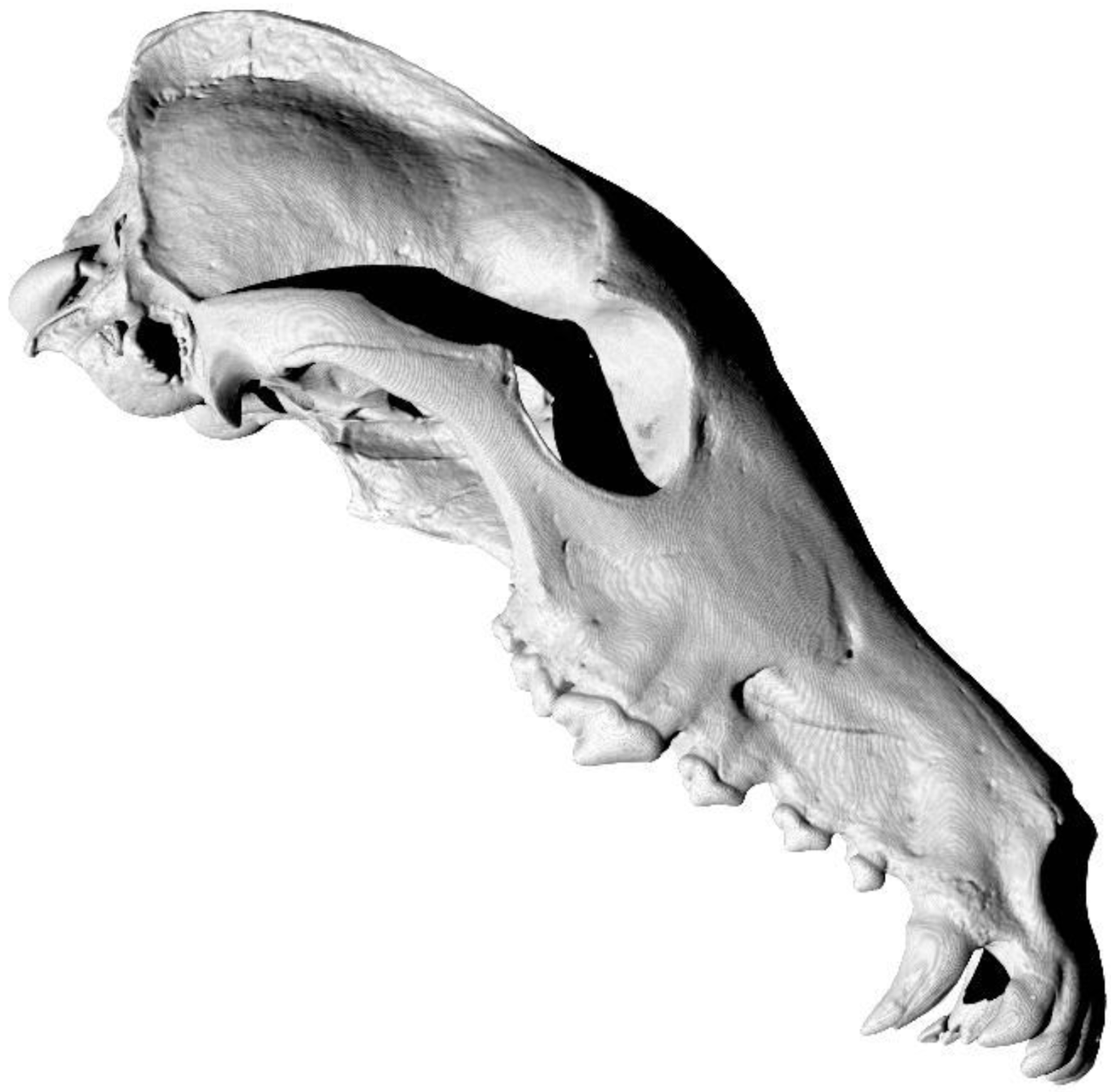
A return to morphology
Questions:
Rules of morphological change
Drivers of morphological change (ecology, historical contingency)
Mechanisms of morphological change (genetic basis)
Unsolved problem: shape
Current state-of-the-art:
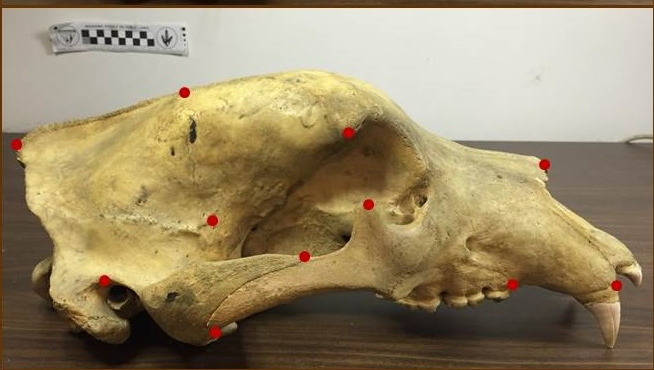
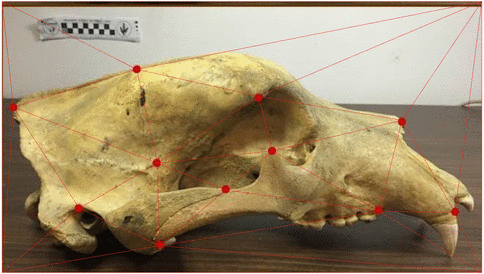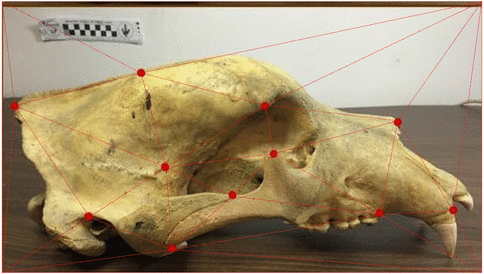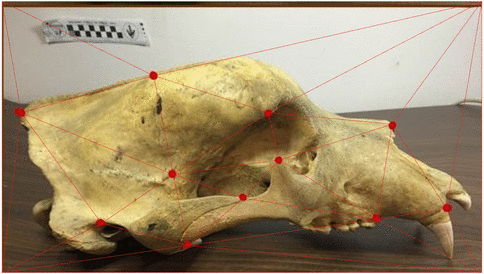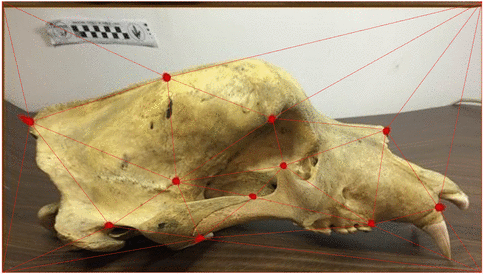
Use of landmarks.
Procrustes alignment.
PCA analyses.
Assume top PCs evolve according to a Brownian motion process.
Then use of methods similar to DNA analyses for computation.
Challenges:
Loss of information in the use of landmarks
Loss of information in linearization of nonlinear shape space
Loss of information in only using the top PCs
Lack of biological interpretability and justification.
Lack of modeling flexibility.
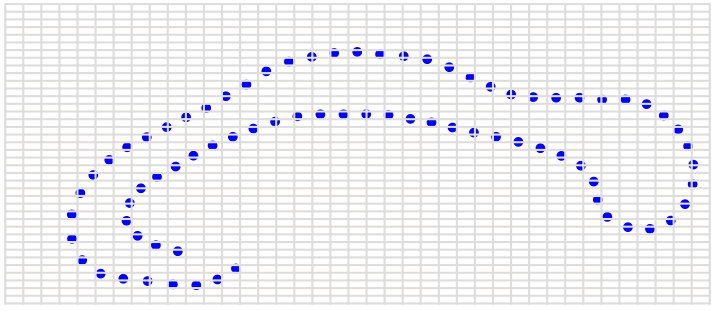
State-of-the-art: full shape is reduced to selected landmarks
Unsolved problem: shape
State-of-the-art: We have high-res digital morphology images but we cannot use the full shape information
- How to model evolution of shape?
- What is the state-space?
- What probability distribution governs the rules of change?
How we do it: Shape analysis
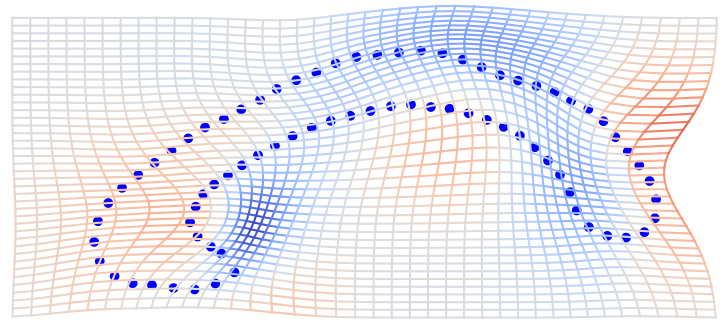
\( \phi \) warp of domain \(\Omega\) (2D or 3D space)
landmarks: \(s=(x_1,\ldots,x_n)\) curves: \(s: \mathbb S^1\to\mathbb R^2\)
surfaces: \(s: \mathbb S^2\to\mathbb R^3\)
\( \phi \)
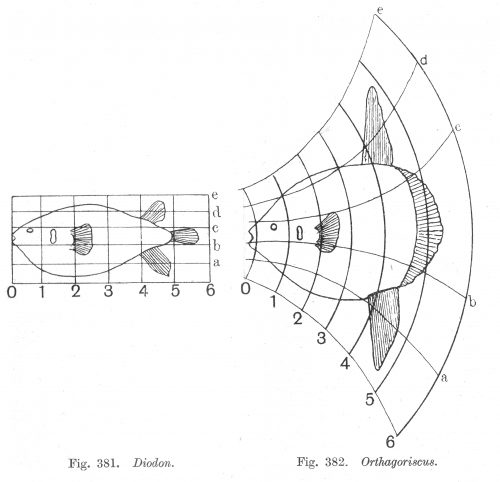
On growth and form, 1917
D'Arcy Thompson
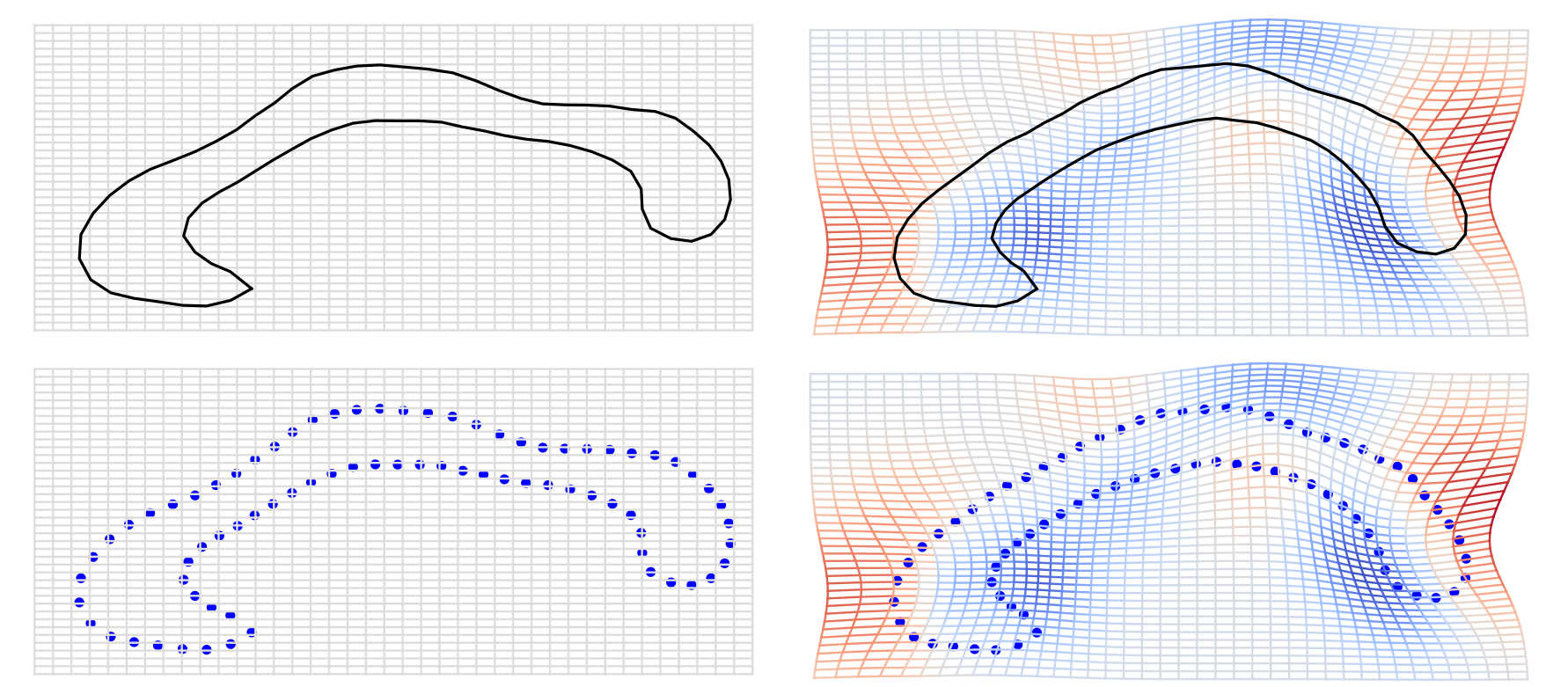

Stochastically evolving shapes
shape \(s_0\)
shape \(s_1\)
stoch. evolution \(s_0\rightarrow s_1\)
Riemannian Brownian motion:
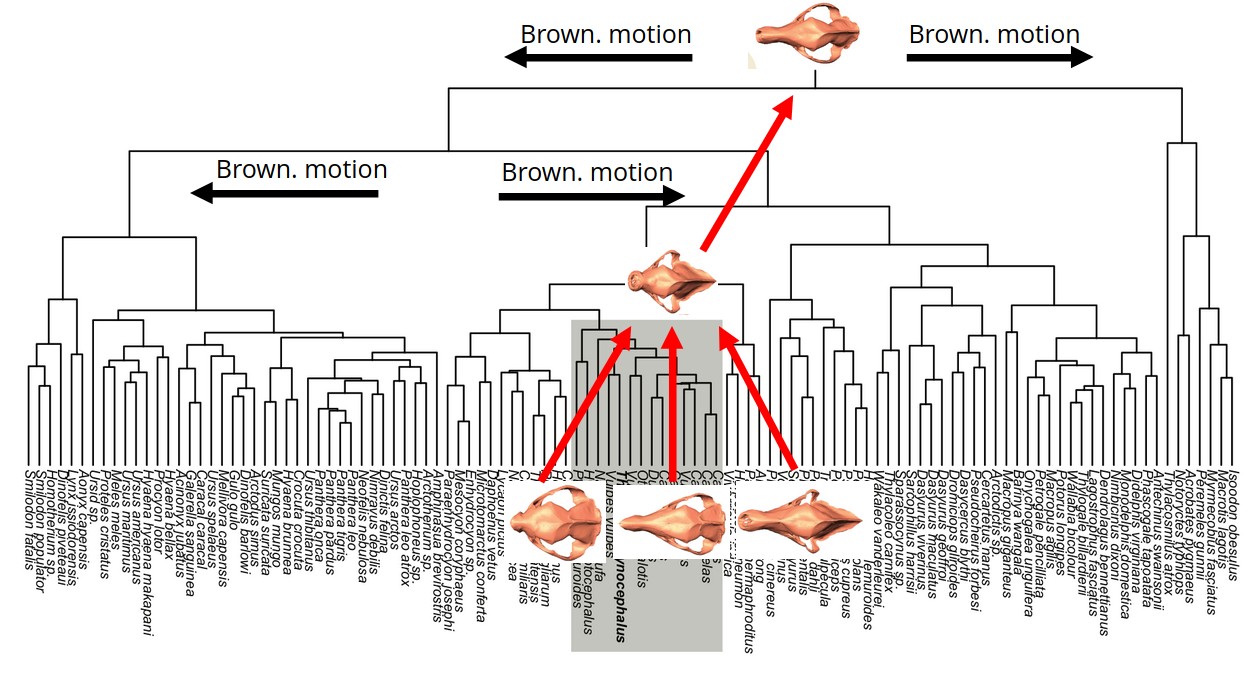
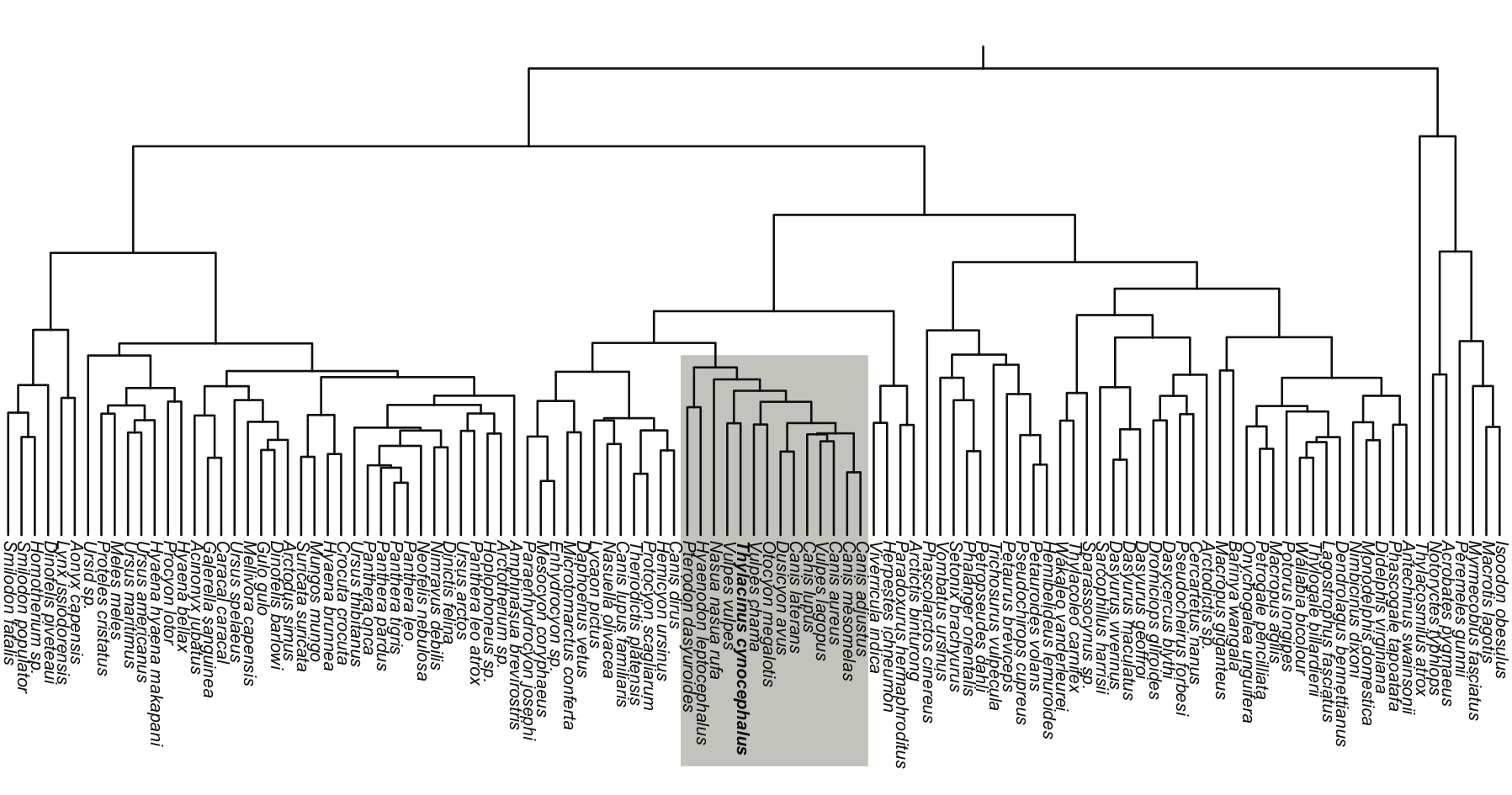
Shapes in phylogenetics
- probabilistic model
- tree pruning for shapes
- stochastic shape matching
- MCMC / variational inference:
- likelihoods
- parameter estimation
- gene/character covariance
- interpolation
- hypothesis testing
- tree inference
inferring the laws of morphological change
Inference: Felsenstein's pruning in modern terms

Brown. motion
Brown. motion
Brown. motion
Brown. motion
branch (independent children)
incorporate leaf observations \(x_{V_T}\) into probabilistic model:
\(p(X_t|x_{V_T})\)
Doob’s h-transform
\(h_s(x)=\prod_{t\in\mathrm{ch(s)}}h_{s\to t}(x)\)
conditioned process \(X^*_t\)
approximations \(\tilde{h}\)
guided process \(X^\circ_t\)
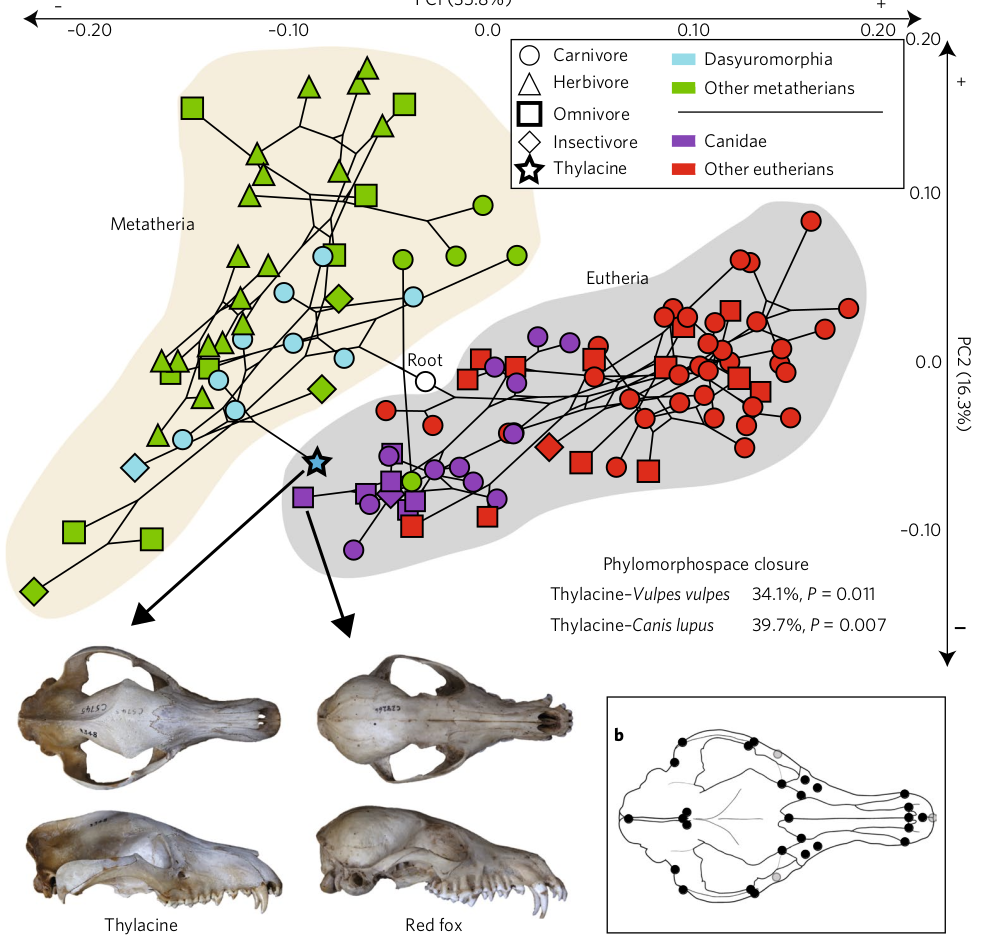
Impact across biology:
Fluctuating asymmetry in butterflies
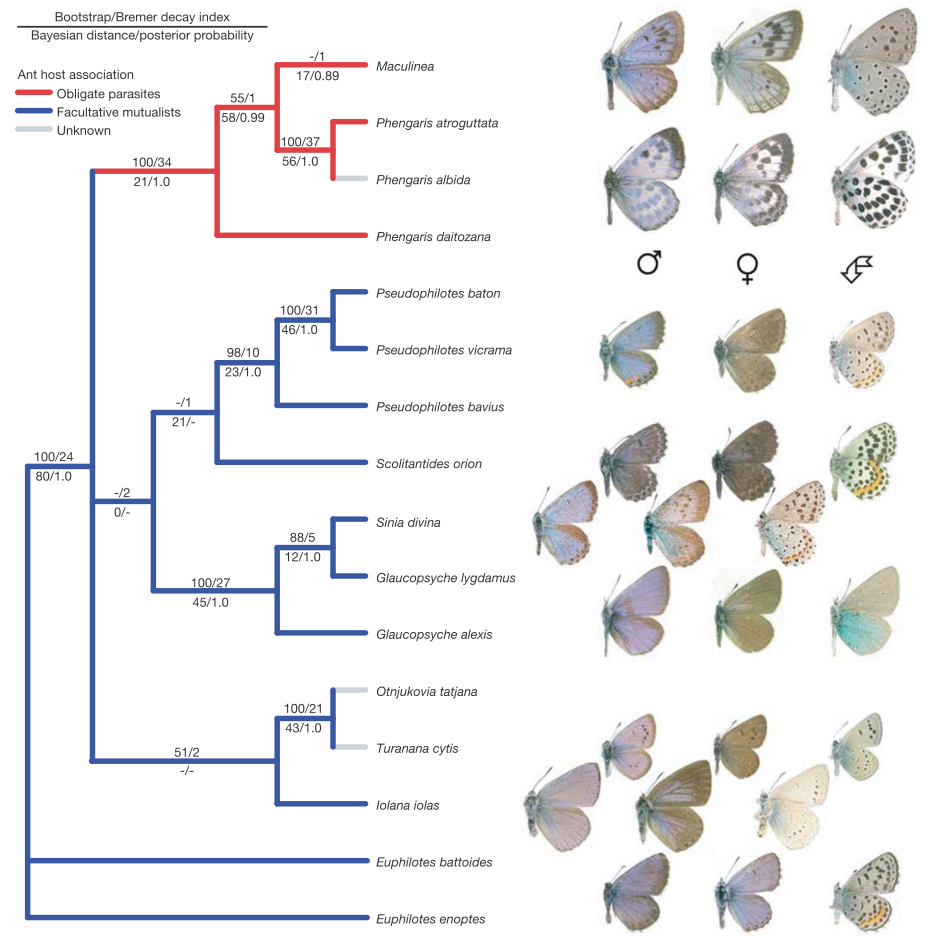
4 cases: Phylogenetics and morphology is foundation for much of bioscience

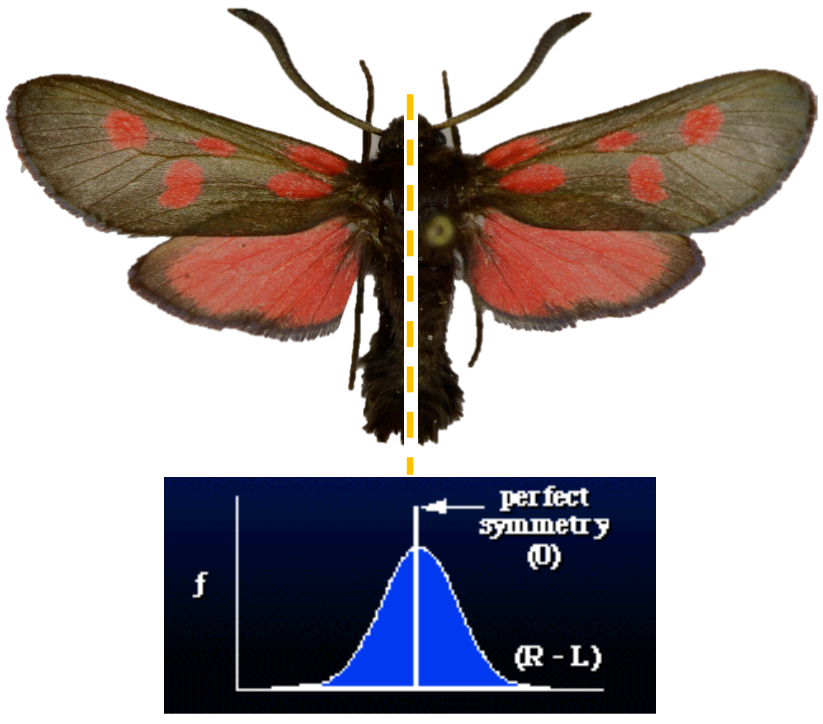
Impact across biology:
Test hypotheses on bird beak evolution genotype/phenotype associations
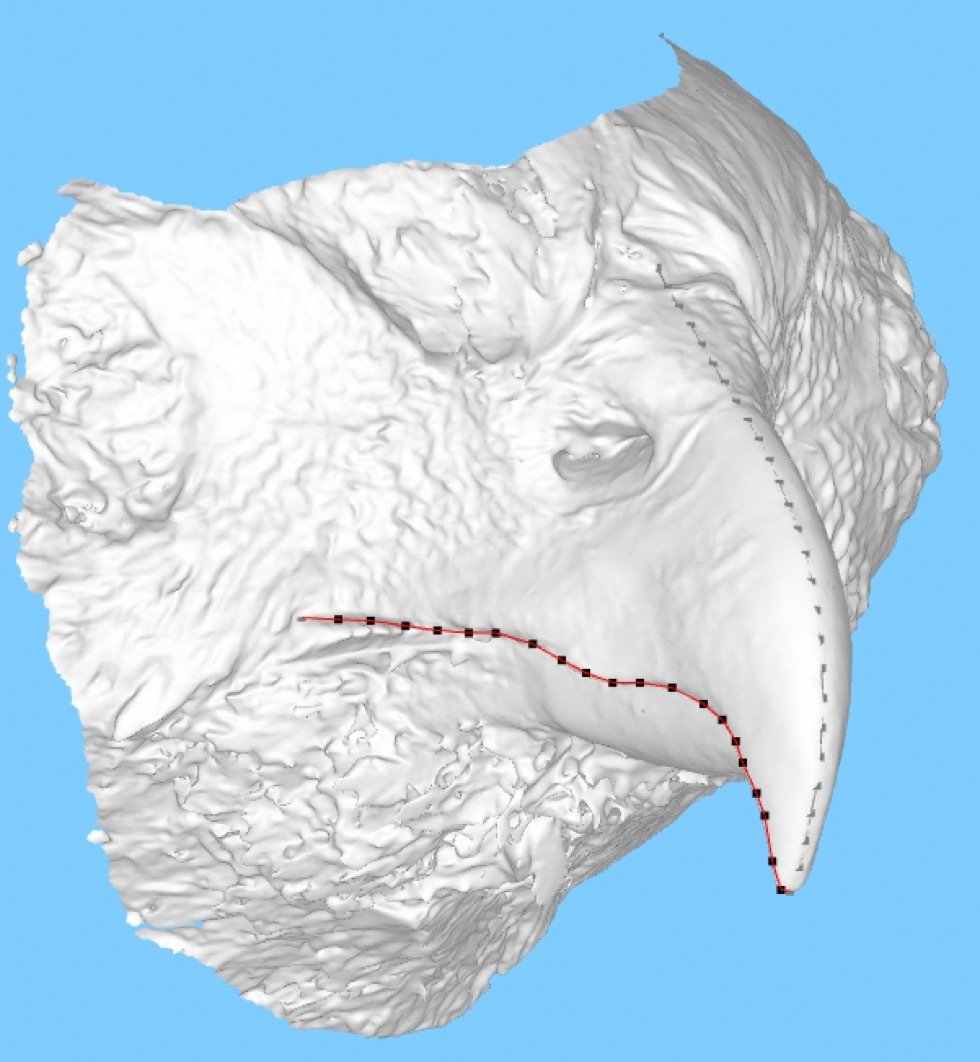
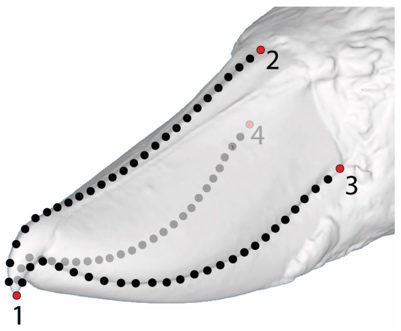
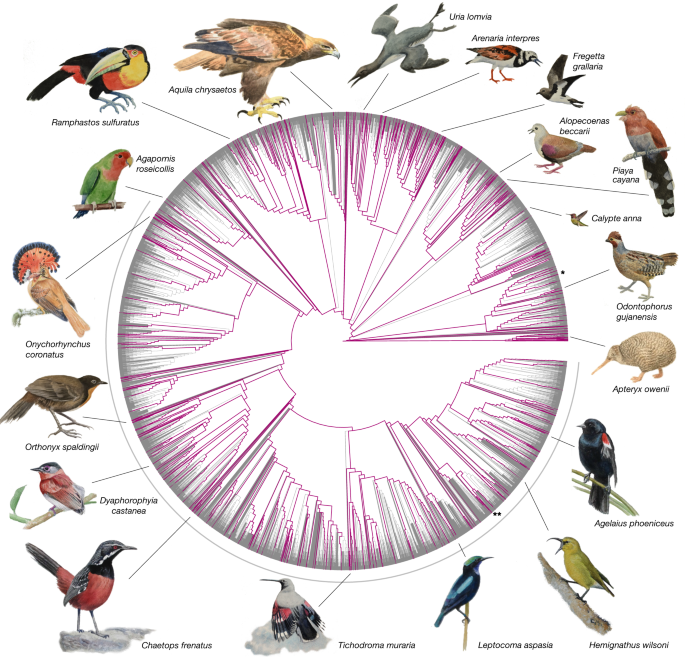
Impact across biology:
Canid evolution and dog domestication
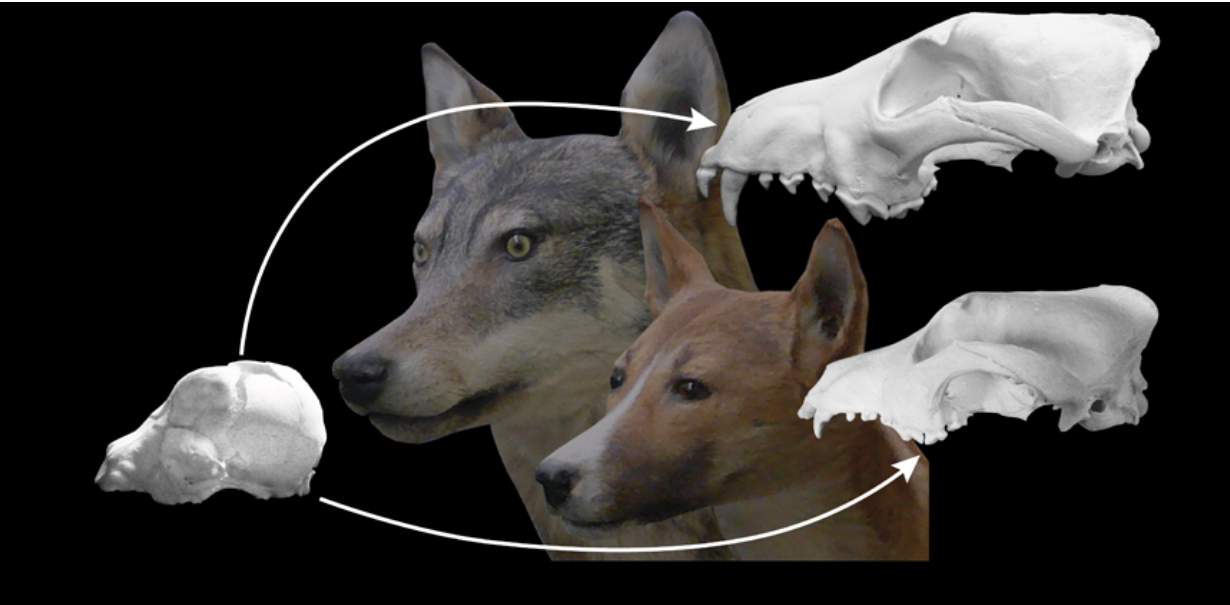


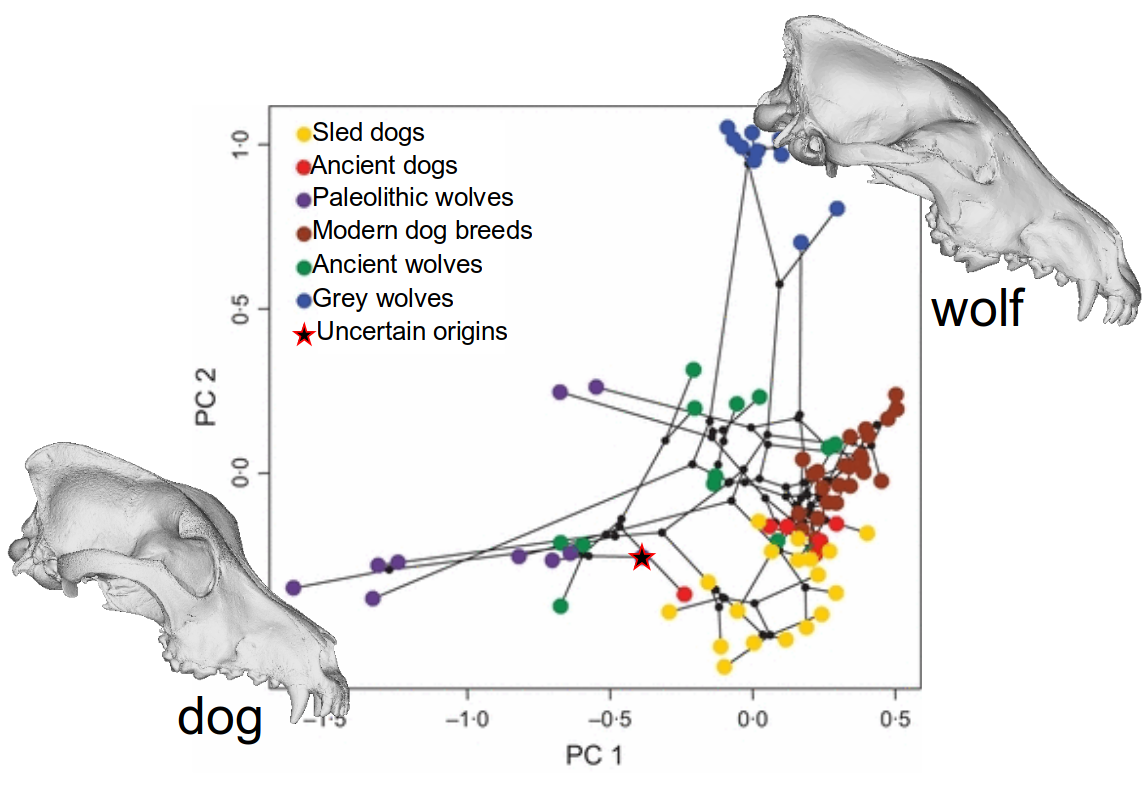
Impact across biology:
Refine palaeohominid tree
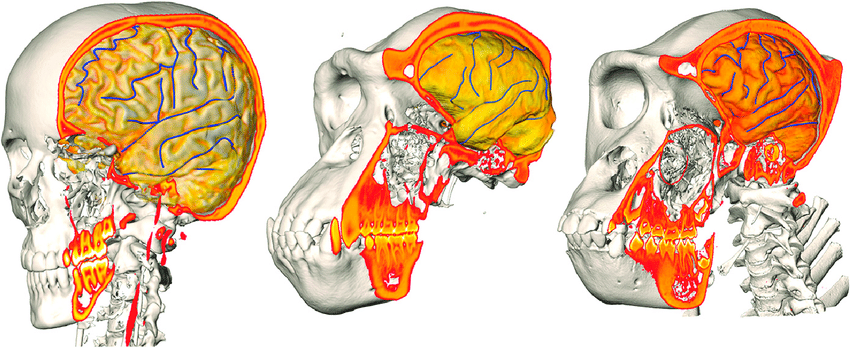

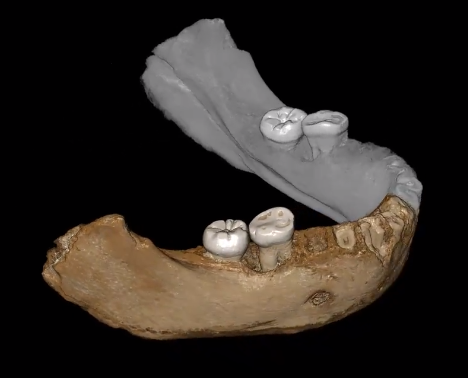
Synergy between phylogenetics, shape analysis, morphology and image analysis
Rasmus Nielsen
GLOBE UCPH, Berkeley
Phylogenetics


Stefan Sommer
DIKU UCPH
Shape analysis
Christy Hipsley
BIO UCPH
Morphology
Mads Nielsen
DIKU UCPH
Image analysis


Collaborators:
Anders Jordan
Natural History Museum of Denmark
Tom Gilbert
GLOBE
Frido Welker
GLOBE
Guojie Zhang
UCPH BIO
Elizabeth Baker, Sofia Stroustrup, Marcus Teller, Lili Bao, Gefan Yang, Liwei Hu, Michael Severinsen, Chao Zhang,
Christine Sarah Andersen + more to come
Josefin Stiller
UCPH BIO