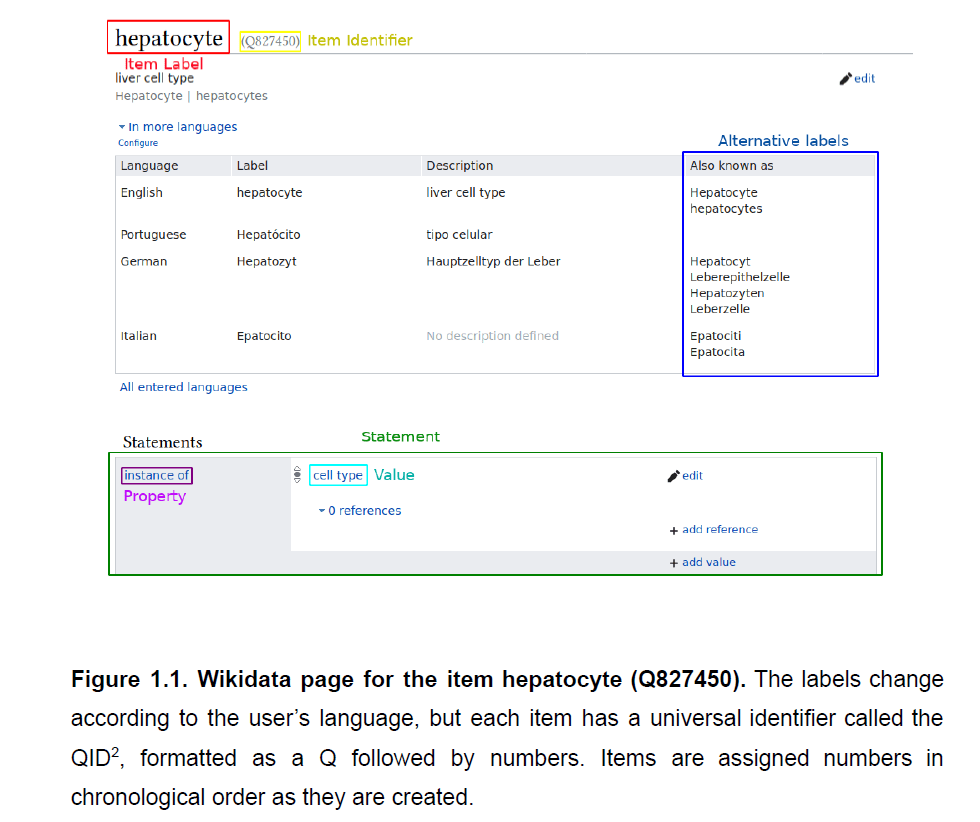
The knowledge graph of Wikidata in the context of the Human Cell Atlas
Student: Tiago Lubiana
Advisor: Helder Nakaya
PhD Thesis Defense - 09/09/2024



Introduction
Motivation

primary sources +
books and reviews
isolated, with ambiguities and inconsistencies
Introduction
Motivation

how do we connect the minds of life scientists?
how do we avoid ambiguity ?
is there a way to make knowledge machine actionable?
how do we leverage computers to reason upon the body of knowledge?
Introduction

Introduction
Wikidata in 2019 was already a tool for the biomedical sciences



Introduction
At the same time, the Human Cell Atlas Project was gaining traction to characterize all existing human cell types




Introduction
What if we leveraged Wikidata to support the Human Cell Atlas?


Introduction
What if we leveraged Wikidata to support the Human Cell Atlas?

Q1. How can Wikidata support bioinformatics research?
Q2. How to use Wikidata to represent knowledge about cell types?
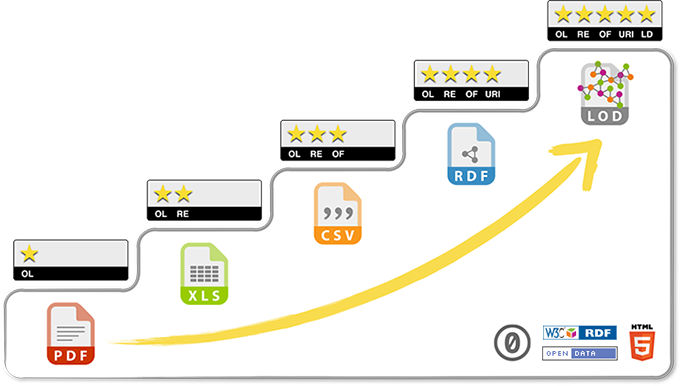
By providing a home for 5-star Linked Open Bio Data
Q1. How can Wikidata support bioinformatics research?

https://5stardata.info/en/




https://5stardata.info/en/



By providing a home for 5-star Linked Open Bio Data
Q1. How can Wikidata support bioinformatics research?
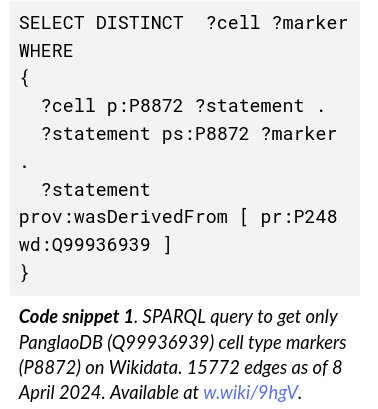
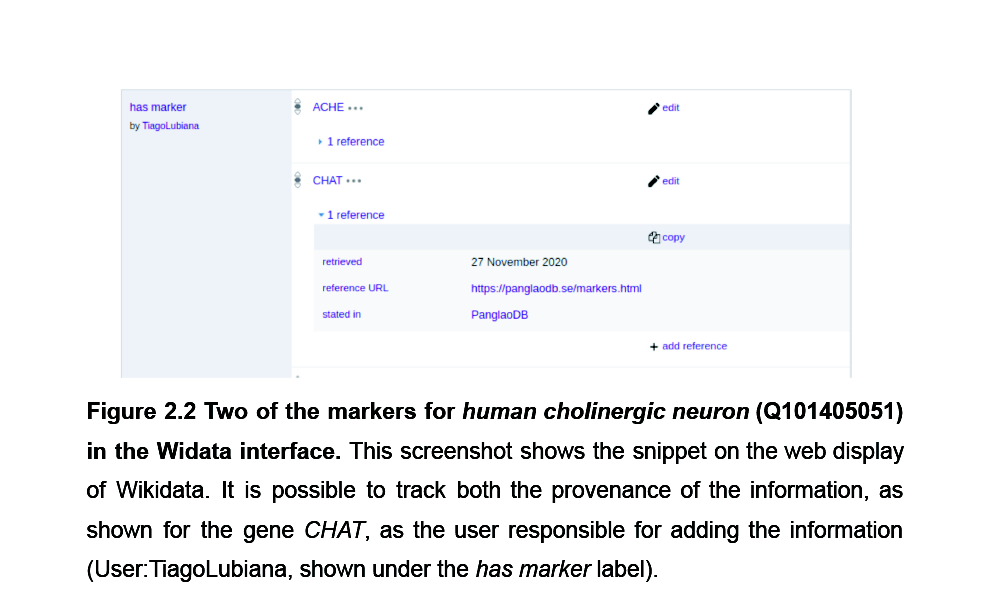
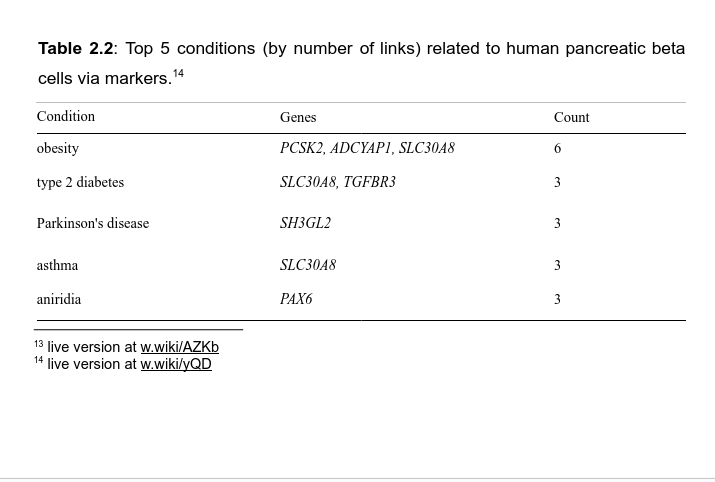
We connected 15.000 cell-gene markers associations from PanglaoDB to Wikidata, making it 5-star Linked Open Data
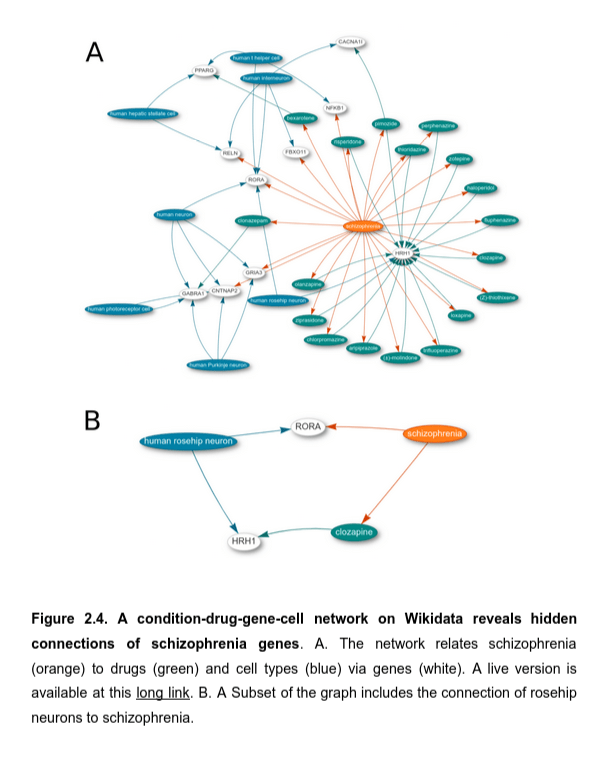
The network structure of Wikidata enables live navigation in a connected knowledge graph, connecting to other pieces of knowledge
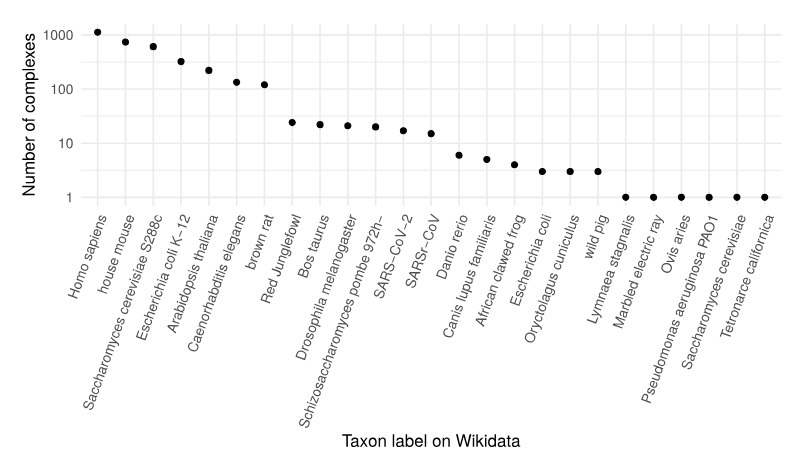
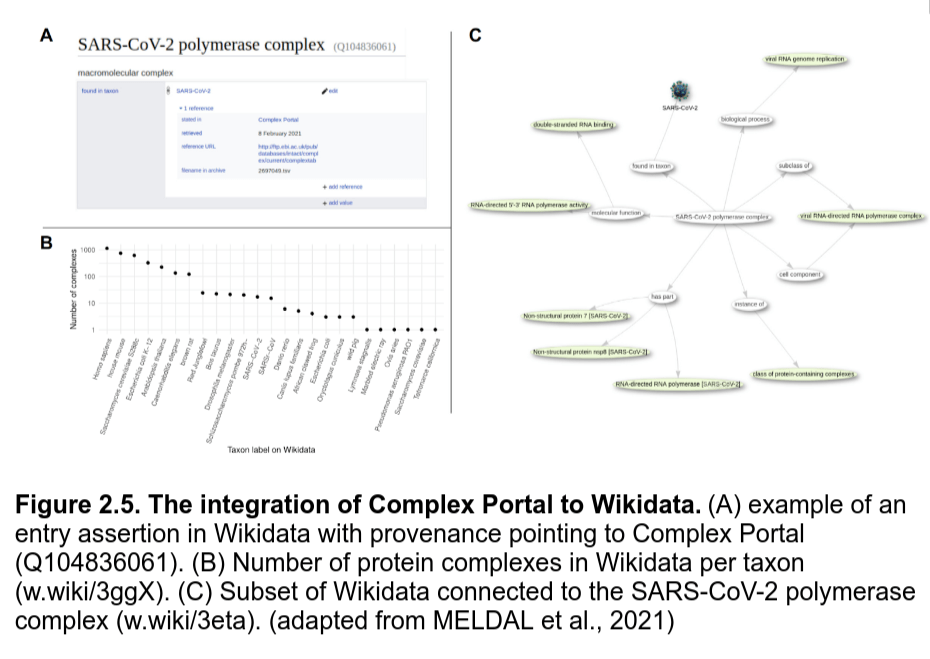
Besides PanglaoDB, we partnered up with the Complex Portal database to make knowledge about complexes 5-star LOD

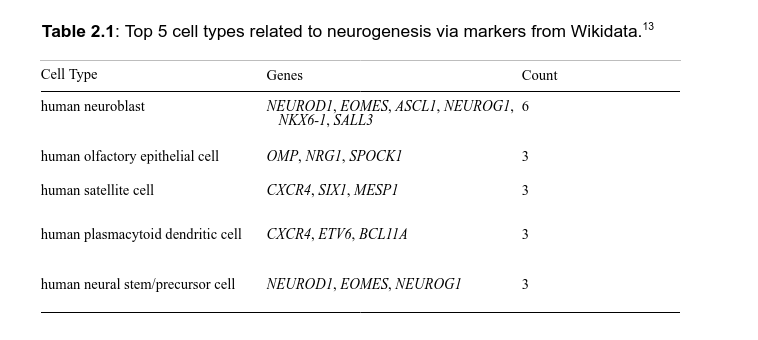
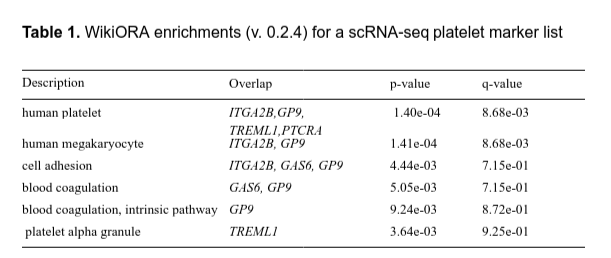
Leveraging SPARQL queries and Wikipedia links for enrichment analysis
What can we do with 5-star Linked Open Bio Data?
Leveraging SPARQL queries and Wikipedia links for enrichment analysis
What can we do with 5-star Linked Open Data?
Leveraging SPARQL queries and Wikipedia links for enrichment analysis
What can we do with 5-star Linked Open Data?
Q2. How to use Wikidata to represent knowledge about cell types?
Where does Wikidata stand in the bio knowledge ecosystem

knowledge portals



data repositories



ontologies and standard vocabularies
rely on





Q2. How to use Wikidata to represent knowledge about cell types?
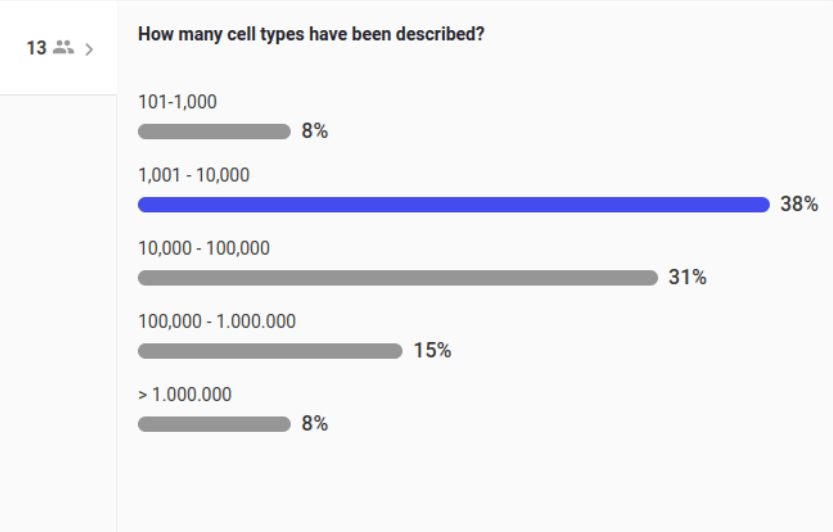
There is a fairly complete list of cell types ... right?

2700

200

80 000 000 000

411

6000

"There is no estimate." (2023)
Aviv Regev
Head of the Human Cell Atlas
Q2. How to use Wikidata to represent knowledge about cell types?
cell types --> cell classes
any idea grouping cells that is used to communicate knowledge about the real world, provided that:
1. it has a (published) name
2. it is useful for theories
3. it is found in multiple individuals, across time
classes
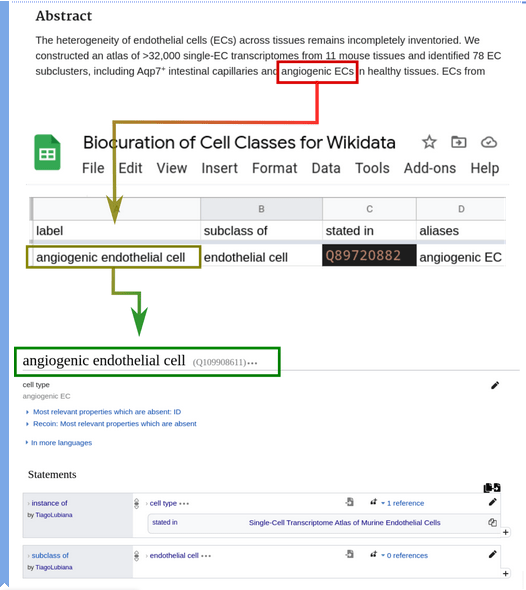
Wikidata for biocuration
- Open edits
- 5-star LOD
- GUI + APIs
- flexible data model
- stable funding
Q2. How to use Wikidata to represent knowledge about cell classes?
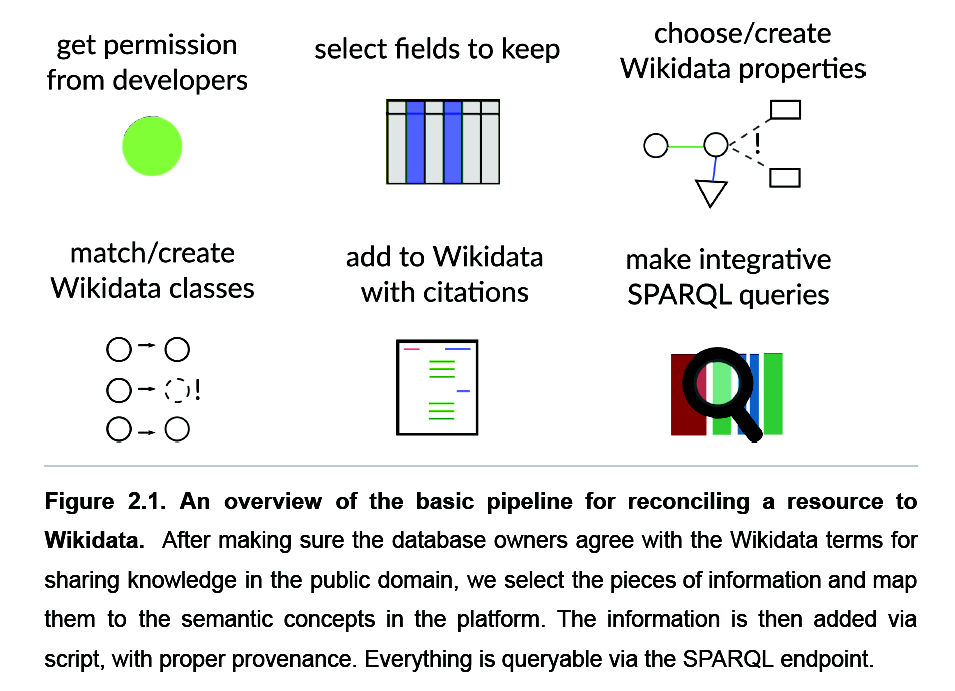
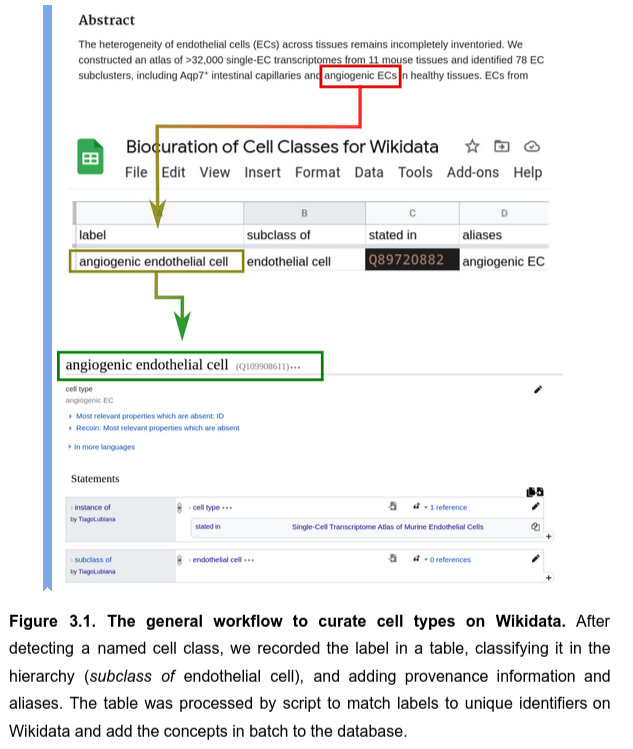
A spreadsheet-based workflow to extract and map information
- read papers related to the Human Cell Atlas
- select cell classes
- curate in a spreadsheet
- parse into Wikidata with a Python script, mapping to unique identifiers
Q2. How to use Wikidata to represent knowledge about cell classes?
Q2. How to use Wikidata to represent knowledge about cell classes?
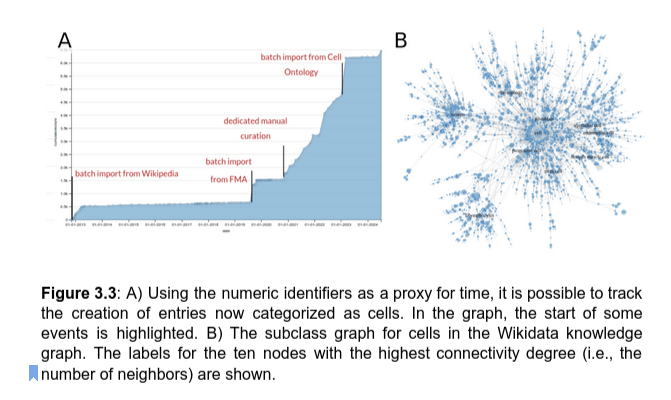
batch import from Wikipedia
batch import
from FMA
dedicated manual curation
batch import from Cell Ontology
Q2. How to use Wikidata to represent knowledge about cell classes?
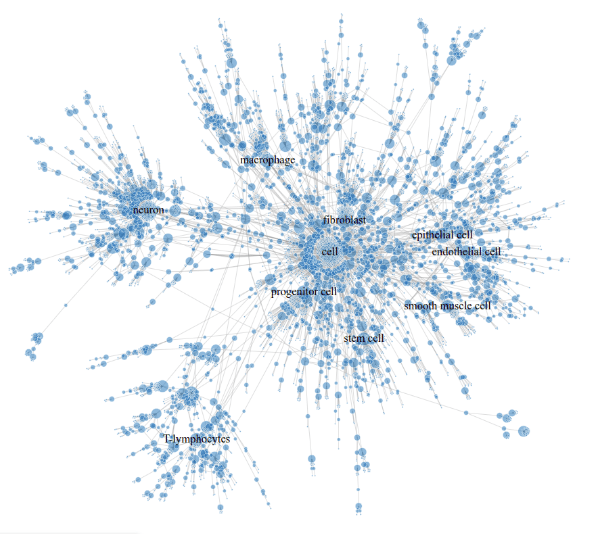
6211 multispecies cell classes - the largest catalog available
Q2. How to use Wikidata to represent knowledge about cell classes?
5837 have at least one supporting reference (w.wiki/AYnP)
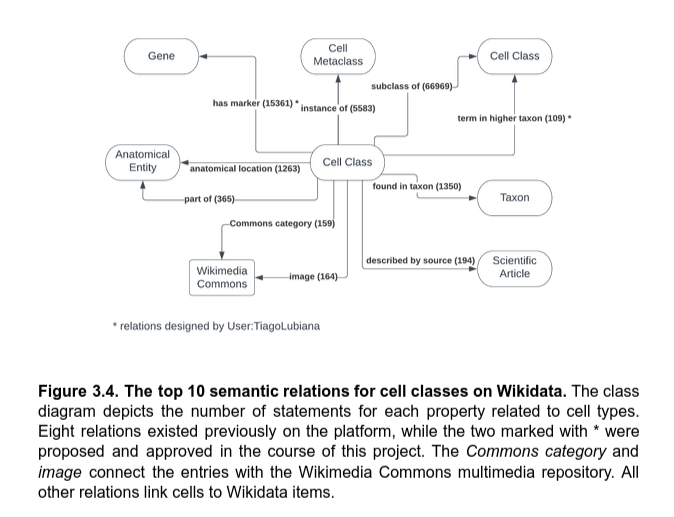
Wikidata as a versatile platform for cell information

- 84 semantic relations
- 150 resources for external identifiers
- links to Wikipedia in 196 languages
Q2. How to use Wikidata to represent knowledge about cell classes?

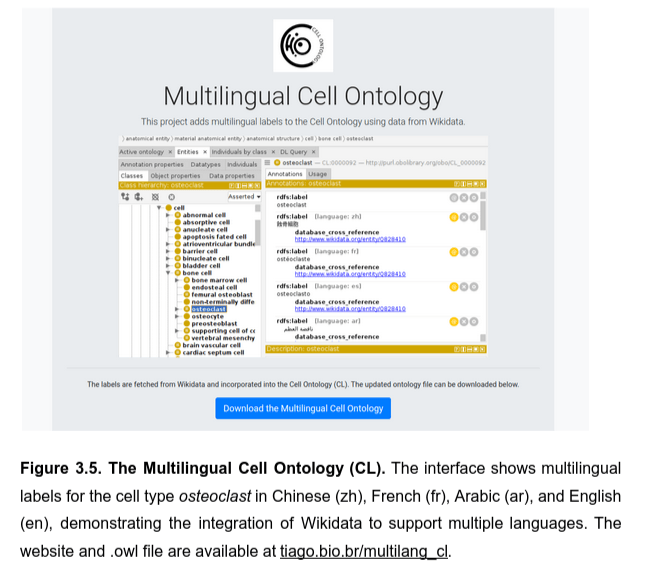
Quickstarting internationalized ontologies
Resurfacing the knowledge about cell classes from Wikidata
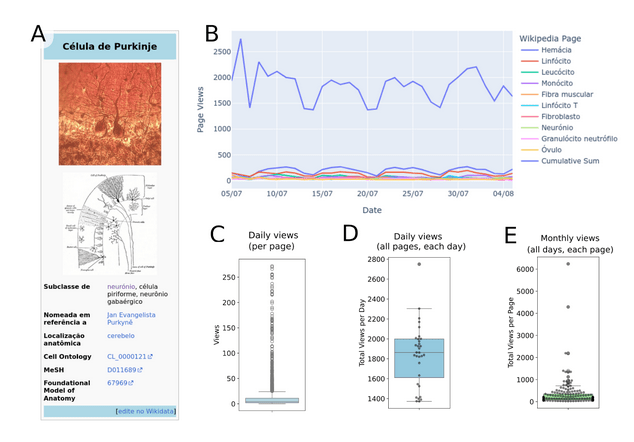
Feeding Wikipedia infoboxes
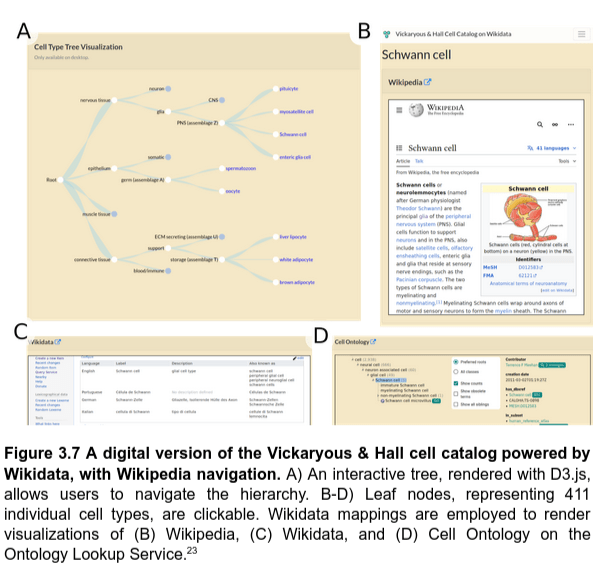
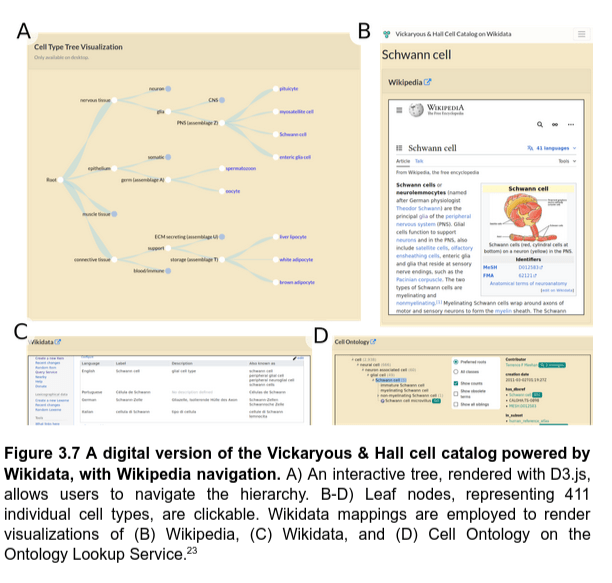
Resurfacing the knowledge about cell types from Wikidata
Powering knowledge portals about cell types

Q2. How to use Wikidata to represent knowledge about cell types?
Conclusion
enriches
custom portals


life sciences research
supports
supports

flexible, fast, multilang decentralized biocuration
crowd curation
data reconciliation

flexible, fast, multilang decentralized biocuration
core resources


standards for

ontologies
complements

enrich
Acknowledgements











The knowledge graph of Wikidata in the context of the Human Cell Atlas
Student: Tiago Lubiana
Advisor: Helder Nakaya
PhD Thesis Defense - 09/09/2024



The knowledge graph of Wikidata in the context of the Human Cell Atlas
Student: Tiago Lubiana
Advisor: Helder Nakaya
PhD Thesis Defense - 09/09/2024









Publications during the PhD
-
Lubiana, T. and Nakaya, H.I., 2024. A reasonable request for true data sharing. The Lancet Regional Health–Americas, 35.
-
Lubiana, T., Lopes, R., Medeiros, P., Silva, J. C., Goncalves, A. N. A., Maracaja-Coutinho, V., & Nakaya, H. I., 2023. Ten quick tips for harnessing the power of ChatGPT in computational biology. PLOS Computational Biology, 19(8), e1011319.
-
Carneiro, C.F.D., da Costa, G.G., Neves, K., Abreu, M.B., Tan, P.B., Rayêe, D., Boos, F.Z., Andrejew, R., Lubiana, T., Malički, M. and Amaral, O.B., 2023. Characterization of comments about bioRxiv and medRxiv preprints. JAMA Network Open, 6(8), pp.e2331410-e2331410
Publications during the PhD
-
Shafee, T., Mietchen, D., Lubiana, T., Jemielniak, D. and Waagmeester, A., 2023. Ten quick tips for editing Wikidata. PLOS Computational Biology, 19(7), p.e1011235.
-
Meldal, B.H., Perfetto, L., Combe, C., Lubiana, T., Ferreira Cavalcante, J.V., Bye-A-Jee, H., Waagmeester, A., Del-Toro, N., Shrivastava, A., Barrera, E. and Wong, E., 2022. Complex Portal 2022: new curation frontiers. Nucleic acids research, 50(D1), pp.D578-D586.
-
Turki, H., Hadj Taieb, M.A., Shafee, T., Lubiana, T., Jemielniak, D., Aouicha, M.B., Labra Gayo, J.E., Youngstrom, E.A., Banat, M.A., Das, D. and Mietchen, D., 2022. Representing COVID-19 information in collaborative knowledge graphs: The case of Wikidata. Semantic Web, 13(2), pp.233-264.
Publications during the PhD
Kilpatrick, A.M., Rahman, F., Anjum, A.,[...], Lubiana, T., [...] Astroz, Y.C., Douglas, J.M. and Eranti, P., 2022. Characterizing domain-specific open educational resources by linking ISCB Communities of Special Interest to Wikipedia. Bioinformatics, 38(Supplement_1), pp.i19-i27.
Rando, H.M., MacLean, A.L., Lee, [...], Lubianat, T., [...] Dziak, J.J., Shinholster, L. and D’Agostino McGowan, L., 2021. Pathogenesis, symptomatology, and transmission of SARS-CoV-2 through analysis of viral genomics and structure. MSystems, 6(5), pp.10-1128.
Lüscher-Dias, T., Dalmolin, R.J.S., de Paiva Amaral, P., Alves, T.L., Schuch, V., Franco, G.R. and Nakaya, H.I., 2022. The evolution of knowledge on genes associated with human diseases. Iscience, 25(1)
Publications during the PhD
-
Turki, H., Jemielniak, D., Taieb, M.A.H., Gayo, J.E.L., Aouicha, M.B., Banat, M.A., Shafee, T., Prud’hommeaux, E., Lubiana, T., Das, D. and Mietchen, D., 2022. Using logical constraints to validate statistical information about disease outbreaks in collaborative knowledge graphs: the case of COVID-19 epidemiology in Wikidata. PeerJ Computer Science, 8, p.e1085.
-
Hoyt, C.T., Balk, M., Callahan, T.J., Domingo-Fernández, D., Haendel, M.A., Hegde, H.B., Himmelstein, D.S., Karis, K., Kunze, J., Lubiana, T. and Matentzoglu, N., 2022. Unifying the identification of biomedical entities with the Bioregistry. Scientific data, 9(1), p.714.
Participation in Events
Organize knowledge in machine readable format
Biocuration is the field of life sciences dedicated to organizing biomedical data, information and knowledge into structured formats, such as spreadsheets, tables and knowledge graphs.

We all know, use, and love biocurated resources

knowledge curation
data curation




Kegg, Reactome, Flybase, Gene Cards...
These resources are, though, hosted mostly in U.S. / Europe.
Previous experience w/ network analysis in CSBL
What could I do as a PhD student in Brazil?
Enter knowlege graphs and Wikidata
Wikidata
* all purpose, openly editable KG
* wealth of biomedical information
* many intersections of academia
I'd love to work with that!