Deep Learning
for QSAR
rl403[at]cam.ac.uk

About Me
- My name is Rich :)
- 3rd year PhD student studying Chemistry at Cambridge University




- My PI is Dr. Andreas Bender
- We have ~20 PhD students and ~5 postdocs
- Work on many cheminformatics and bioinformatics problems, including
- Gene expression data analysis
- Drug repurposing and repositioning
- Drug toxicity modelling
- Mode of action analysis
- Biological network analysis
- Drug combination modelling
- Collaborate widely with industry and academia, and growing quickly!


PyData
- I learned to program 3 years ago
- I mainly use 3 languages:
- Python evangelist!



Deep Learning for QSAR
Deep Learning for QSAR
It's unreasonably good
- Image recognition
- Speech recognition
- NLP
- and many more...
Automatic Captioning

Show, Attend and Tell: Neural Image Caption Generation with Visual Attention, Xu et al.
| arXiv:1502.03044 |

Deep Learning for QSAR
Deep Learning for QSAR
Quantitative Structure Activity Relationship
The basic assumption for all molecule based hypotheses is that similar molecules have similar activities. This principle is also called [Quantitative] Structure–Activity Relationship ([Q]SAR).
”
(thanks wikipedia)
Deep Learning for QSAR
Quantitative
Regression rather than classification

Deep Learning for QSAR
Structure
atoms

bonds
Caffeine
carbon atom
Deep Learning for QSAR
Activity
Activity is a broad term...
- Protein affinity, inhibition or activation
- Cell toxicity
- Phenotypic responses
- Physical properties (QSPR)
How are these measured?

IC50
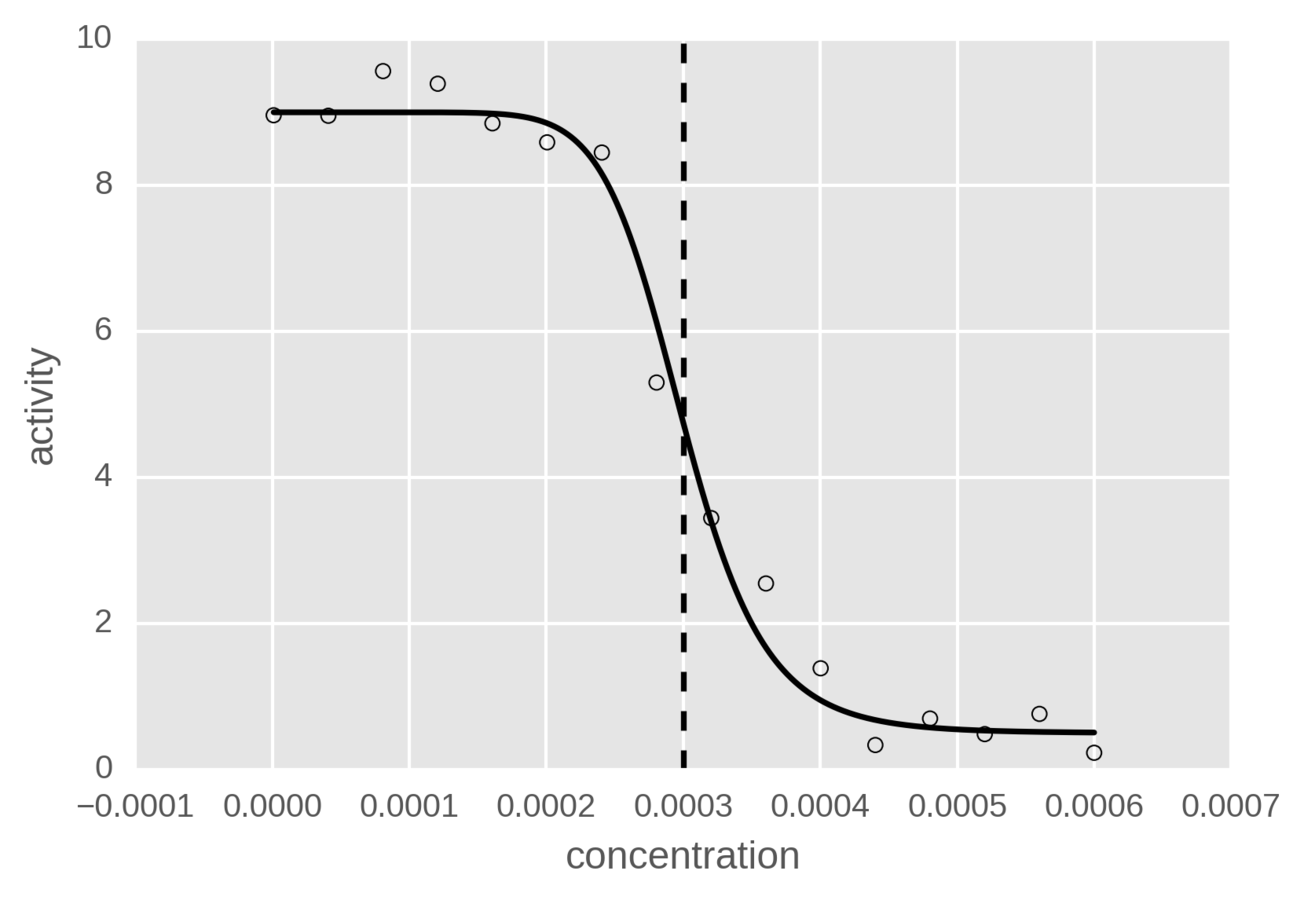
Deep Learning for QSAR
Relationship

7.4
A QSAR model is a mapping between a chemical structure and a number.
This may also include a why as well as a what.
What is it good for?

- Testing new drugs is expensive
- It would be good to virtually screen out compounds that are unlikely to work
- It would also be good to predict the mode of action of a drug
Why should we use deep learning to build QSAR models?
Feature Learning
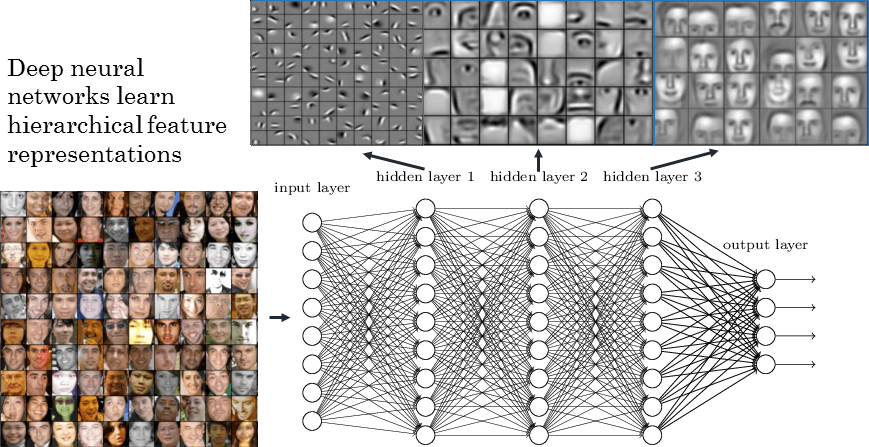

Cat detector
Atoms

Functional Groups
Pharmacophores
Raw pixels
Edges
Facial features
Faces

Increasing complexity


Multitask learning


- Recognising each number is its own task
- Sharing weights between tasks allows statistical power to be shared between tasks




x 1000s
The data
Cheminformatics Data Sucks!
- The data is almost always sparse. Compounds are unlikely to be tested on all outputs.
- The data is usually very noisy. Errors are often on the scale of an order of magnitude!
- Data is sometimes wrong. A compound can interfere with how an assay works, giving false readings.
- Data is often unevenly distributed. Many compounds tend to be derived from a common scaffold.
- Data is inconsistent. Even for the same task, data is usually from different assays, measured using different techniques.
ChEMBL
- ChEMBL collects open access data.
- Cleans the data.
- Links to standard identifiers.
- Provided in a relational format:



- 250 000 compounds
- 500 000 activities
- 710 proteins
- 0.24% density


number of compounds per protein
Data Summary
Featurization

0: C
1: C, N, N
2: C, C, C, C, N
8201
5
Repeat for all atoms!
Circular Fingerprints
hash
modulo-2048

1
1
The Modelling


Classification Model
from keras.models import Sequential
from keras.layers import Dense, Dropout, BatchNormalization
from keras.regularizers import WeightRegularizer
from keras.optimizers import Adam
model = Sequential([
Dense(2000, input_dim=2048, init='he_normal', activation='relu'),
BatchNormalization(),
Dropout(0.5),
Dense(2000, init='he_normal', activation='relu'),
BatchNormalization(),
Dropout(0.5),
Dense(581, init='he_normal', activation='sigmoid')])
model.compile(optimizer=Adam(lr=0.0005), loss='binary_crossentropy')Binary crossentropy:

hist = model.fit(X_train, Y_train,
nb_epoch=250, batch_size=2048,
class_weight=Y_train.sum(axis=0),
callbacks=[ModelCheckpoint('classification.h5')],
validation_data=(X_valid, Y_valid))Training



precision: 0.6243
recall: 0.6702
f1: 0.6465
mcc: 0.6464
roc_auc: 0.9785
pr_auc: 0.6654

Comparison to other techniques
Did it really work?
- Caffeine is in the test set, so the model has never seen it before.
- Let us try to predict for it:
>>> caff_pred = pd.Series(model.predict(caff_fp)[0], index=Y.columns)
>>> caff_pred = caff_pred.sort_values(ascending=True)
target_id
P29275 0.809085 # Adenosine receptor a2b
P29274 0.428097 # Adenosine receptor a2a
P21397 0.295609 # Monoamine oxidase A
P27338 0.241524 # Monoamine oxidase B
P33765 0.076975 # Adenosine receptor A3
dtype: float64
Identifying features


Extracting information from a black box, using a black box!

0 0
1 0
2 0
3 1
4 0
..
2045 0
2046 1
2047 0
Name: paclitaxel, dtype: uint8
0 0
1 0
2 0
3 0
4 0
..
2045 0
2046 1
2047 0
Name: caffeine, dtype: uint8
0.809
0.740
+0.069 difference


Adenosine a2b
Monoamine oxidase
Regression Model
from keras.models import Sequential
from keras.layers import Dense, Dropout, BatchNormalization
from keras.regularizers import WeightRegularizer
from keras.optimizers import Adam
model = Sequential([
Dense(2000, input_dim=2048, init='he_normal', activation='relu'),
Dropout(0.5),
BatchNormalization(),
Dense(2000, init='he_normal', activation='relu'),
Dropout(0.5),
BatchNormalization(),
Dense(710, init='he_normal', activation='linear')])
model.compile(optimizer=Adam(0.0001), loss=sum_abs_error, metrics=[r2])Sum absolute error:
Only one problem...
NaN
Fixing the loss function
K.is_nan = T.isnan # tf.is_nan
K.logical_not = lambda x: 1 - x # tf.logical_not
def sum_abs_error(y_true, y_pred):
valids = K.logical_not(K.is_nan(y_true))
costs_with_nan = K.abs(y_true - y_pred)
costs = K.switch(valids, nan_cost, 0)
return K.sum(costs, axis=-1)
- Missing values in our target variables cause the NaN.
- These should not contribute to the cost (the loss for those targets should be zero).

hist = model.fit(X_train, Y_train,
nb_epoch=250, batch_size=2048,
class_weight=Y_train.sum(axis=0),
callbacks=[ModelCheckpoint('classification.h5')],
validation_data=(X_valid, Y_valid))


train
valid
test

Comparison to other techniques
| Neural Network | Random Forest | |
|---|---|---|
| R2 | 0.722 | 0.528 |
| MSE | 0.546 | 0.707 |
Only for Dopamine D2 - receptor
Estimated error in the data ~0.4, so this is good!
Transfer Learning
model = Sequential([
Dense(2000, input_dim=2048, init='he_normal', activation='relu'),
Dropout(0.5),
BatchNormalization(),
Dense(2000, init='he_normal', activation='relu'),
Dropout(0.5),
BatchNormalization(),
Dense(20, init='he_normal', activation='linear')])
model.compile(optimizer=Adam(0.0001), loss=sum_abs_error)
model.fit(X_train, Y_train[selected_targets])
new_model = Model(inputs=model.inputs[0], outputs=model.layers[-4].output)
new_model.compile('sgd', 'use')
X_deep = new_model.predict(X_train)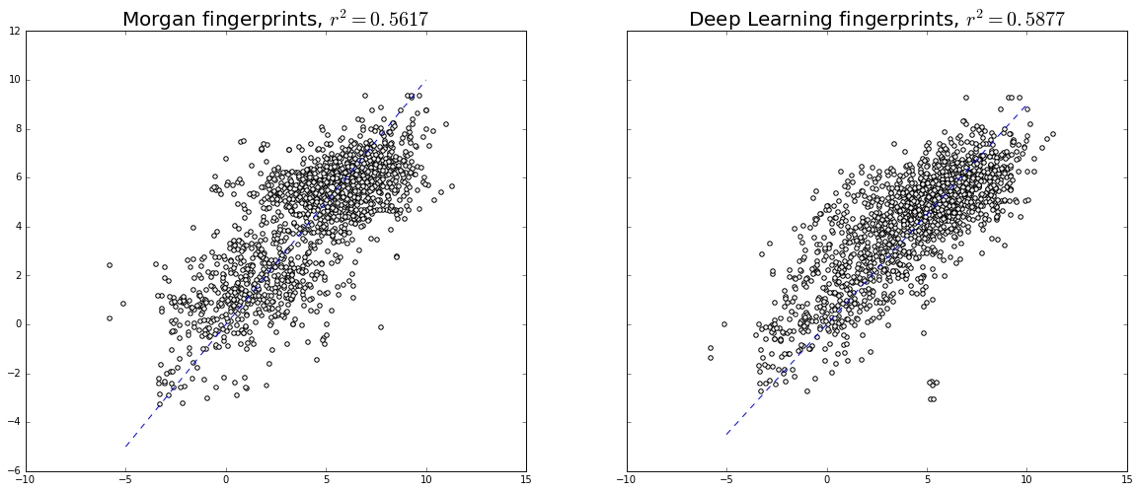
Acknowledgements

Andreas Bender
Günter Klambauer
The Bender group
PyData sponsors




