Opal 2.x
OBiBa’s core database application for biobanks
About this presentation
-
Hit the left/right arrow to browse to the main sections
-
Hit the up/down arrow to see the slides in each section
-
Hit the "escape" key to see all the slides
You can Try it !
-
username: administrator
-
password: password
What's new
- Projects
- Administration
- R
- DataSHIELD
Projects
A project is a repository of data and dictionaries.
In a project, data are imported, exported, analysed, transformed and shared.
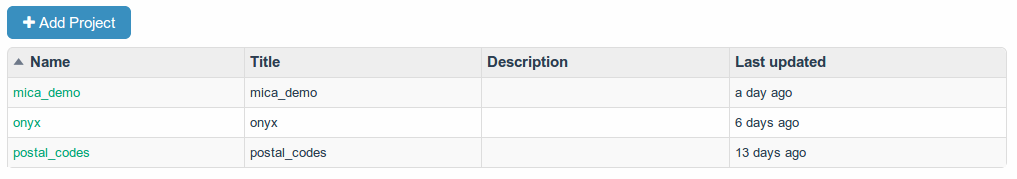
Project Tables
Formerly known as a datasource.

Project Tables
A table has a dictionary (variables) and values.
- Each variable can be edited individually,
-
There is a summary for each value type.

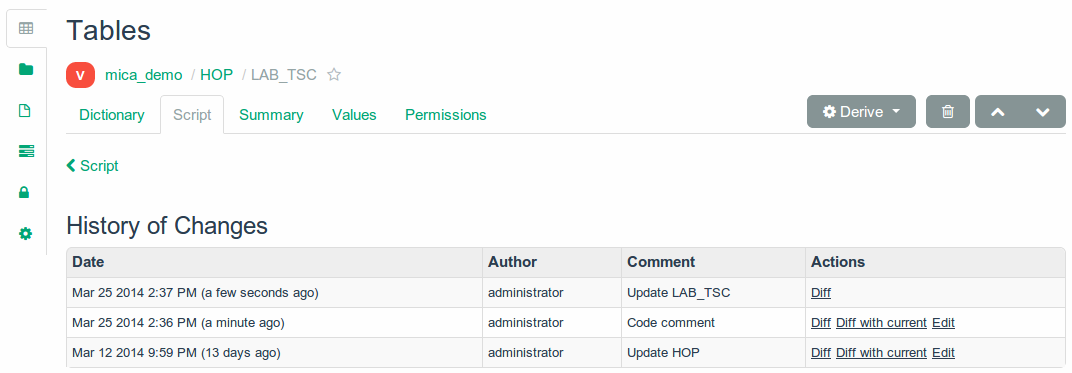
Project Tables
Derived variable scripts versioning.
Project Files
A project has its own folder to allow collaborative work.

Project Reports

Project Tasks
Follow import, export, copy, report tasks.

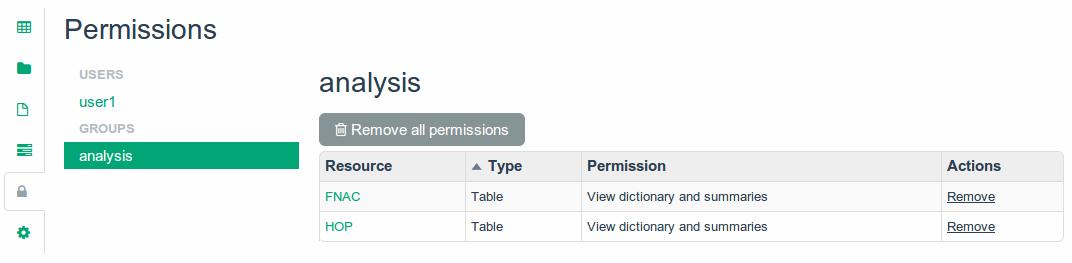
Project Permissions
Manage project permissions in one page.
Project Administration
Description, data storage, key store...

Administration
Administration was improved to make Opal more dynamic.
- Data access: users, ...
- Data management: identifiers, ...
- Data analysis: R, ...
- System: databases, JVM...
Users
-
Users and groups can be added from the web interface,
-
Users can be authenticated by password or certificate.
Identifiers Mappings
-
Identifiers of any entity type can be mapped,
- Easier identifiers management.
R Service
R server can be controlled, even if it runs on a different machine.
Databases
- Easier database management,
-
MongoDB for large datasets.
Java Virtual Machine
-
Memory usage
- Threads
- Garbage collector activity
R
Opal and R in 5 lines.
# Login in opallibrary(opal)o <- opal.login('administrator', 'password', 'http://localhost:8080')# Assign a table to a data.frameopal.assign(o,'D','mica_demo.FNAC')# Analyse data in the remote R sessionopal.execute(o,"summary(D)")# Get data into the local R sessionD <- opal.execute(o,"D")# Terminate remote R sessionopal.logout(o)
DataSHIELD
DataSHIELD process is parallelized.
Was sequential
- R command is executed on Opal 1
- ...
- R command is executed on Opal n
- Results are combined
Now is parallel
- R command is submitted to Opal 1, ..., Opal n
- R command result is retrieved from Opal 1, ..., Opal n
- Results are combined