HLI HG38 DNASeq
Summary
- VCF File Characteristics
- Sample characteristics
- High Pass Regions
- Manifest of IDs
VCF File Characteristics
- N = 2357 files
- N variants (Min,Mean,Max)
- (4,495,629; 4,738,841; 5,846,745)
- Not phased
- Not QCed
Sample characteristics
- N=2357
- Sample volume
- DNA concentration
- DNA mass
- DNA purity 260/280 ratio
| Characteristic | 1-missingness | Mean (SD) |
|---|---|---|
| Sample Volume (uL) | 100% | 60.15 (3.80) |
| DNA mass (ng) - calculated | 99.6% | 13693.10 (10186.93) |
| DNA concentration (ng/uL) | 99.6% | 228.58 (170.61) |
| DNA purity (260/280) | 17.5% | 1.72 (0.10) |
Notes:
- There is negligible protein contamination.
- Min mass >2220 ng, Min volume > 30 uL, Min concentration >27ng/uL
- Ilumina usually requires a minimum of 10 nM in 20 uL
- Min nM>80 assuming insert size of 50bp + 2x 151 bp paired-end nmer
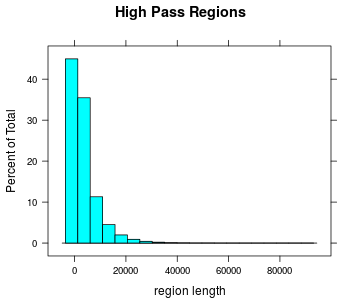
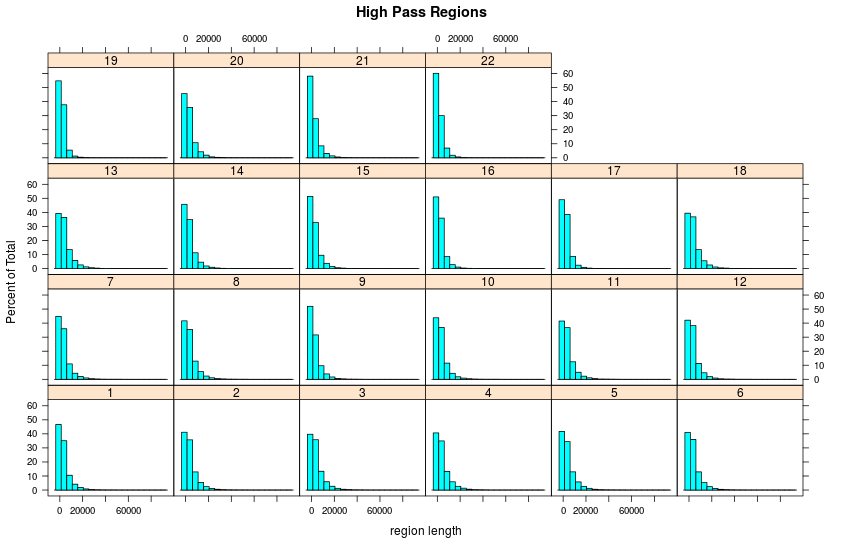
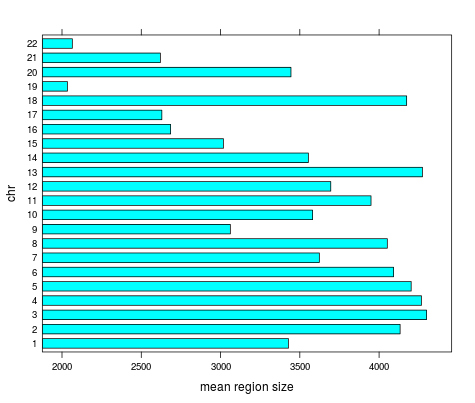
Highpass regions
- Length
- Number
- Total length
Number of regions | chr
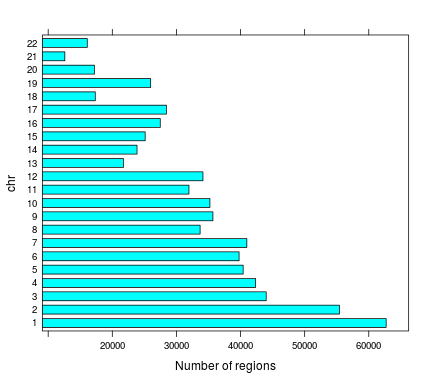
Total number of regions 712,265
Length of highpass regions
Length | chr
Mean length | chr
Highpass total length | chr
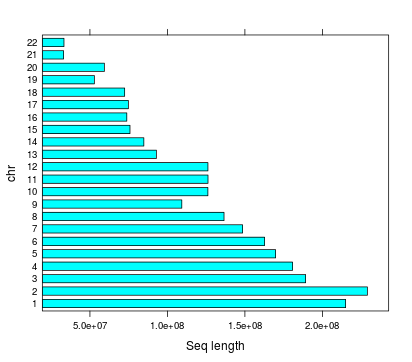
2,572,251,922 (84%)
HG38 Total non-N lenght = 3,074,968,030
Manifest of IDs
- Number of individuals sent
- Number of individuals sequenced
- Number of twin pairs and zygosity
- Number of trios
Analysis of IDs
| Samples | Retrieved | Submitted | Intersection |
|---|---|---|---|
| N | 2,357 | 2,069 | 2,036 |
| Setdiff | 321 | 33 | 0 |
| Gender | 1971/65 | ||
| Singletons | 134 | 134 (46/88) | |
| Twins pairs | 951 | 951 (401/550) | |
| Triplets | 0 | -- | |
| Trios | 159 (with DNA) | 161 (41/120) | |
| Parents | 175 (with DNA) | ||
| Missing from DB | 106 | ||
| Missing Annot | 40 |
Total = Singletons + 2xTwins + Parents = 2211
Omics overlap
| Omics | N |
|---|---|
| PainExomes | 272 |
| GOT2DExomes | 100 |
| UK10K | 861 |
| EB_Fat | 545 |
| EB_Skin | 516 |
| EB_LCL | 586 |
| EB_WB | 298 |
| Fat_450K | 449 |
Meeting 2
10 Aug 2016
Summary
- Summary quality statistics chr20
- Concordance Analysis
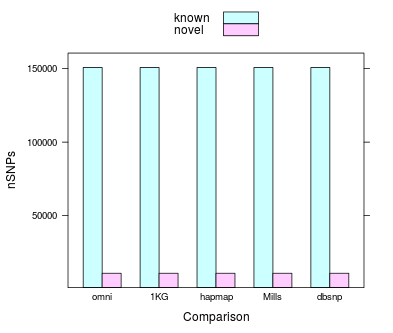
#SNP
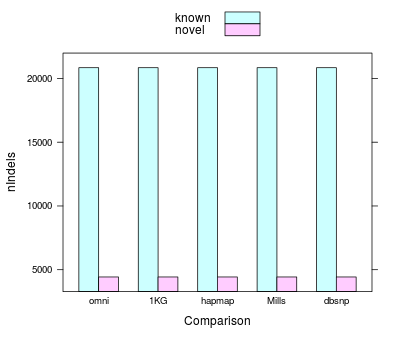
#InDel
#Insert / #Deletion
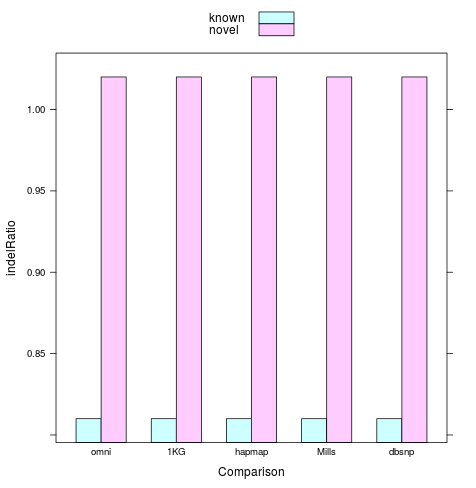
~1.03 in Montgomery 2013 GR
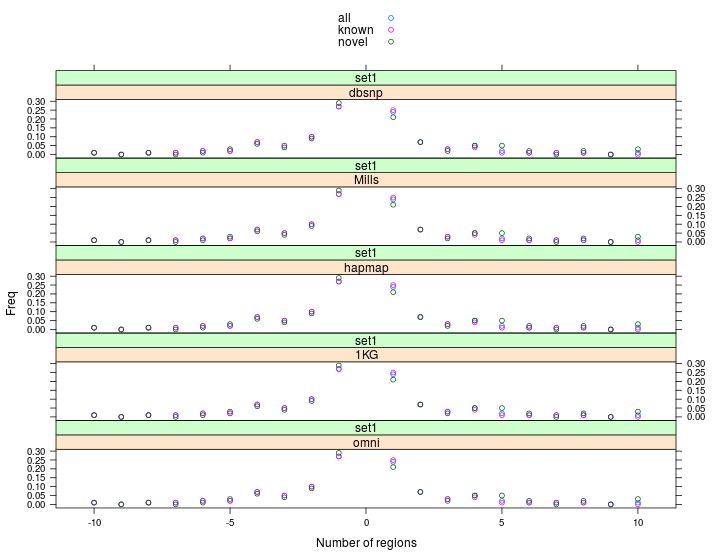
Histogram indels | novelty
Ti/Tv
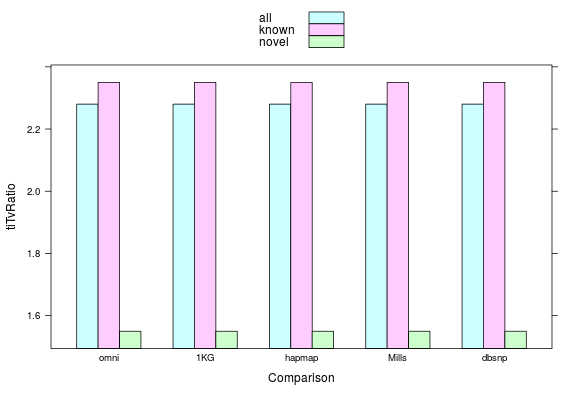
ratio of transition (Ti) to transversion (Tv) ~ 2.0-2.1
Concordance Rate
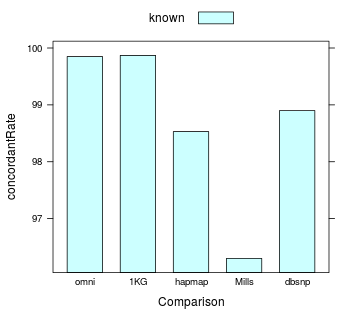
number of concordant sites (that is, for the sites that share the same locus as a variant in the comp track, those that have the same alternate allele) / total
Variant call set FDR
dbSNP
FDR=0.08
FP=14,458
TPhat=168,759
Total=183,804
#MultiIndels / #Indels
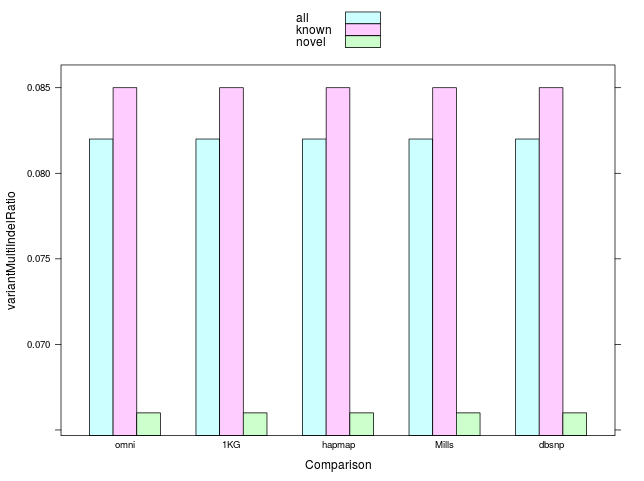
#MultiSNP / #SNP=0.01
Multiallelic variants
Ti/Tv
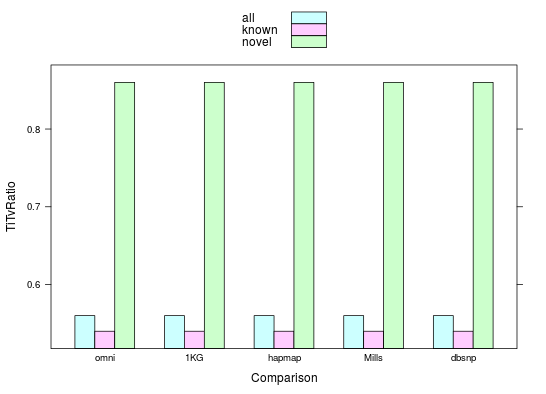
Multiallelic variants
SNP Novelty Rate
knownSNPsPartial- the number of loci at which at least one allele in eval was found in the known comparison file
knownSNPsComplete- the number of loci at which all alleles in eval were also found in the known comparison file
SNPNoveltyRate- the sum of knownSNPsPartial and knownSNPsComplete divided by nMultiSNPs
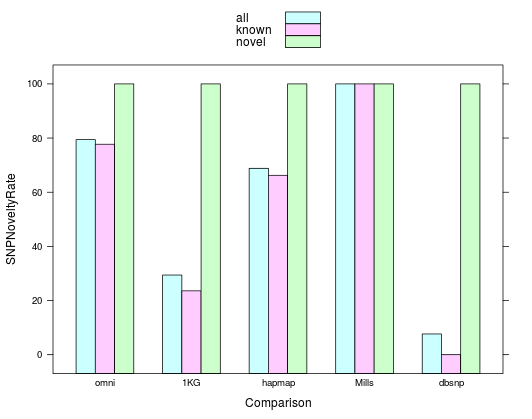
Multiallelic variants
Filter Chr20: vcftools
Merge VCF: GATK CombineVariants/VCFmerge
WG HLI HG38 VCF
Convert to plink: vcftools, GATK
flip +ve strand: plink
Annotate VCF: ANNOVAR
Filter highpass regions: Bedtools
gen file
matrixeqtl file
phasing: shapeit2
GenotypeConcordance: GATK
multiallelic test
Exon HG19 vcf
Coordinate change HG38:
liftOver/crossmap
convert files: qctool / gtools
plink file
phased plink file
HLI MEETING
14 SEP 2016
CHR20 Stats
- P HWE
- P Heterozigozity
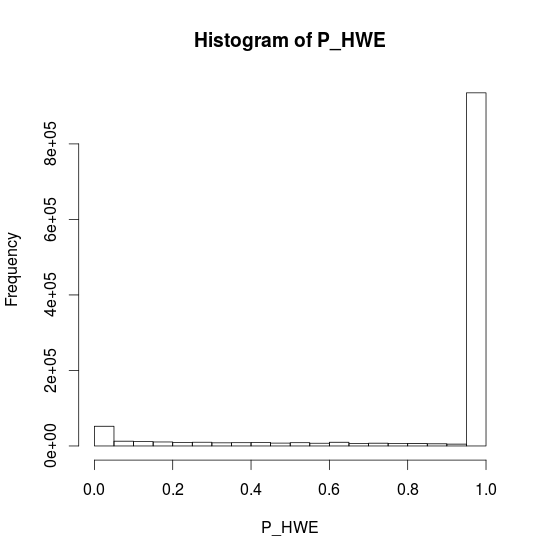
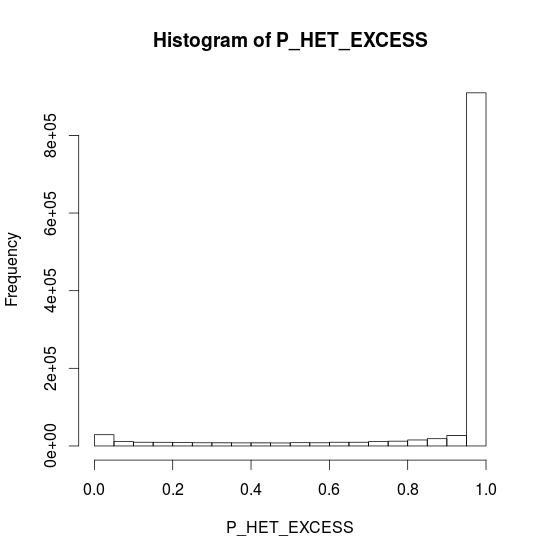
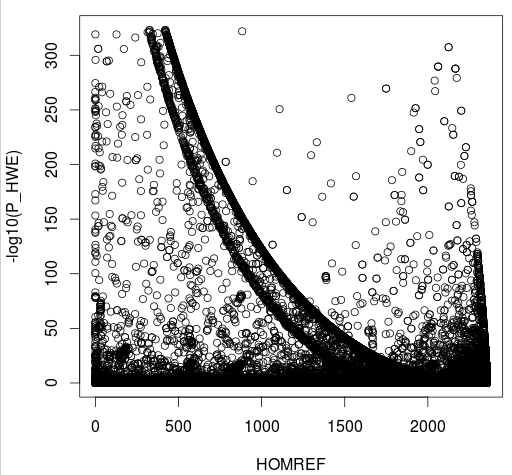
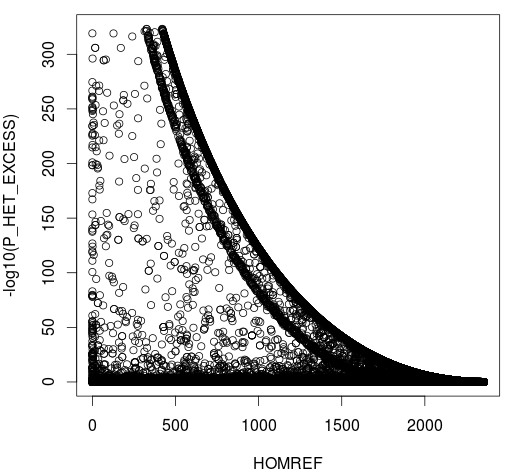
CHR20 Stats
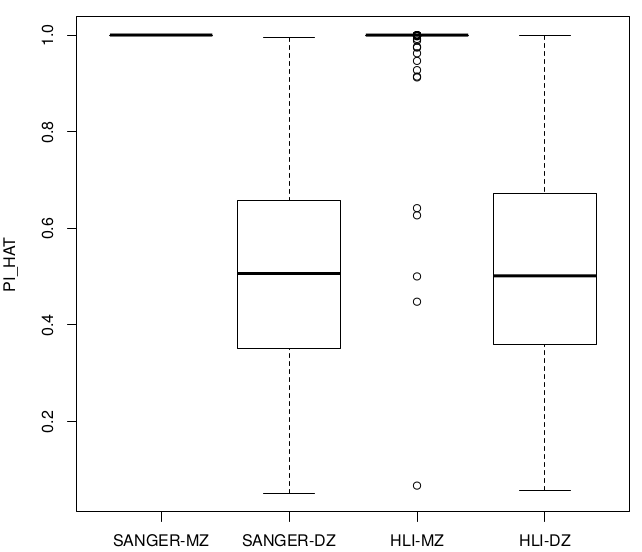
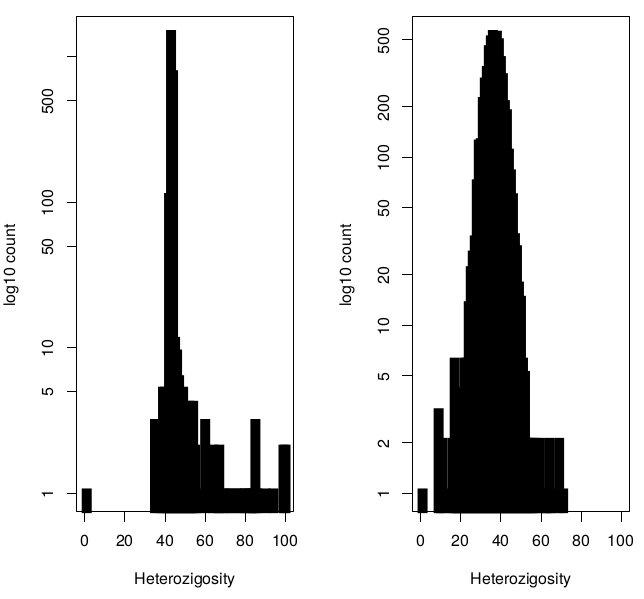
All chr analysis
- Analysis conducted on SNPs shared by HLI WGS and Genotyping arrays
- Comparison of statistics with and without exclusion of individuals with high heterozygosity (Het>0.4)
Exclusions
-
42 individuals excluded due to high (Het>0.4) heterozygosity :
- 5 Twin Pairs (10 individuals)
- 22 unrelated (unpaired twins)
- 10 parents
Discordant individuals
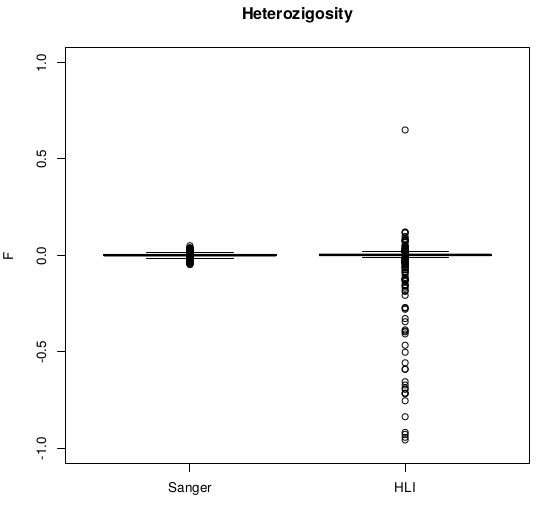
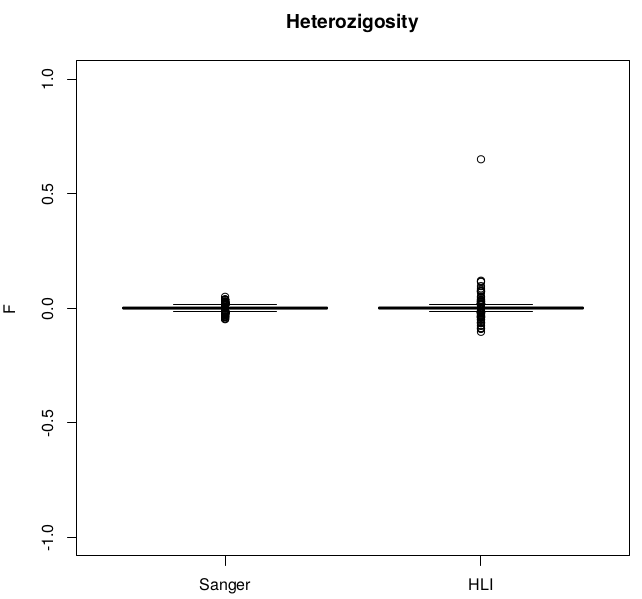
Heterozigosity
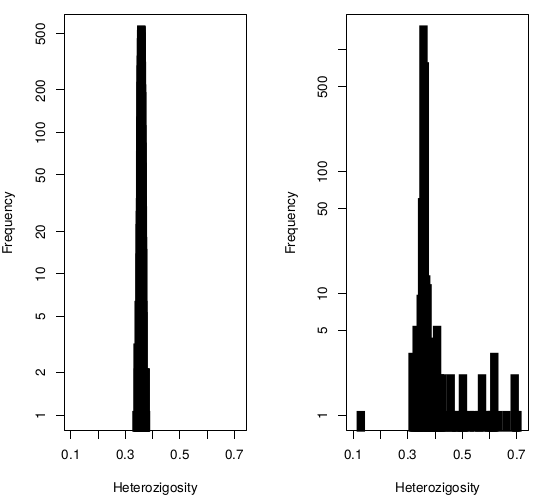
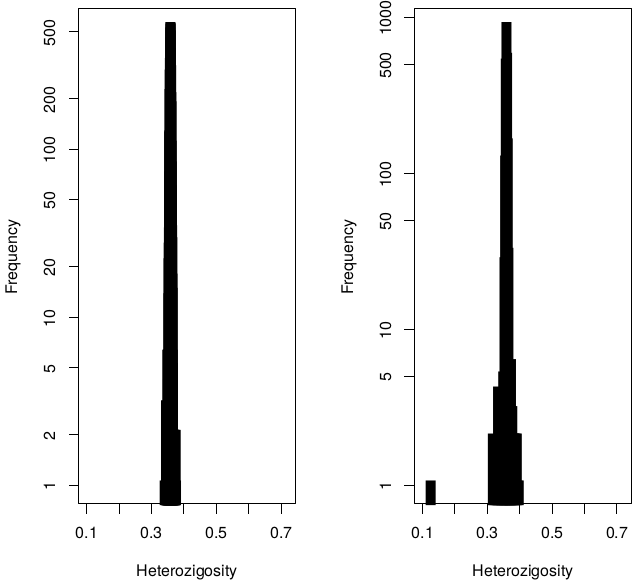
Heterozigosity -- F
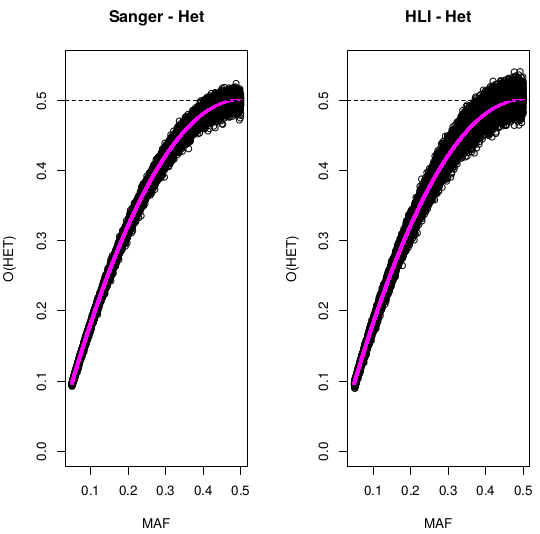
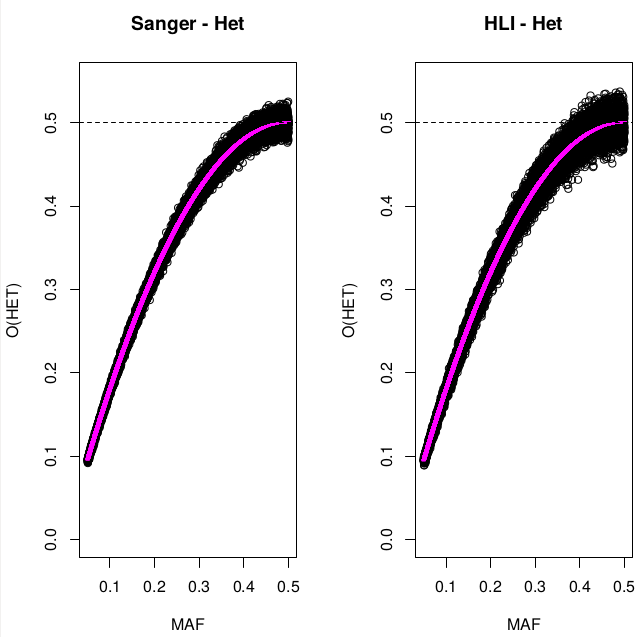
Heterozigosity O&E ~ MAF
Relatedness ~ Zygosity
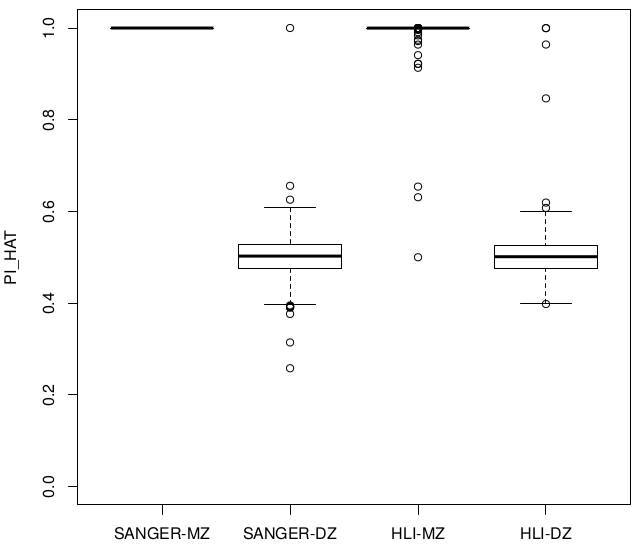
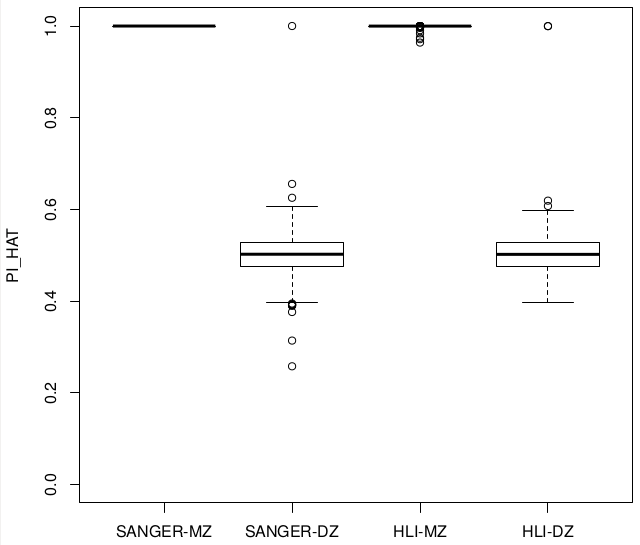
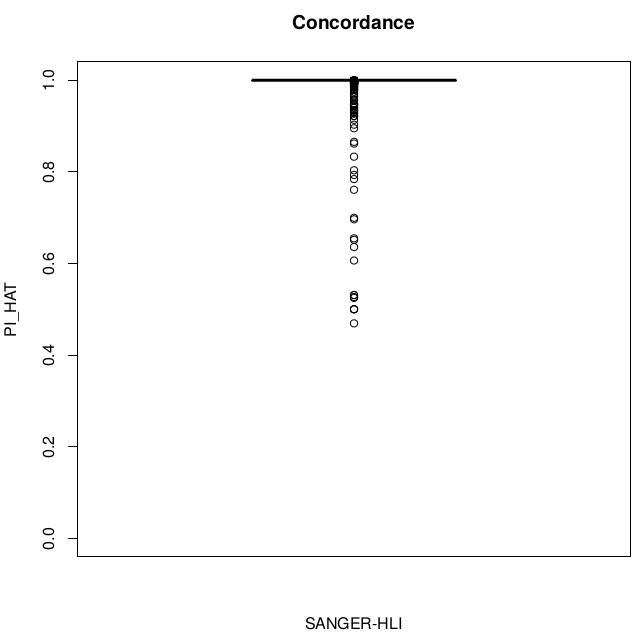
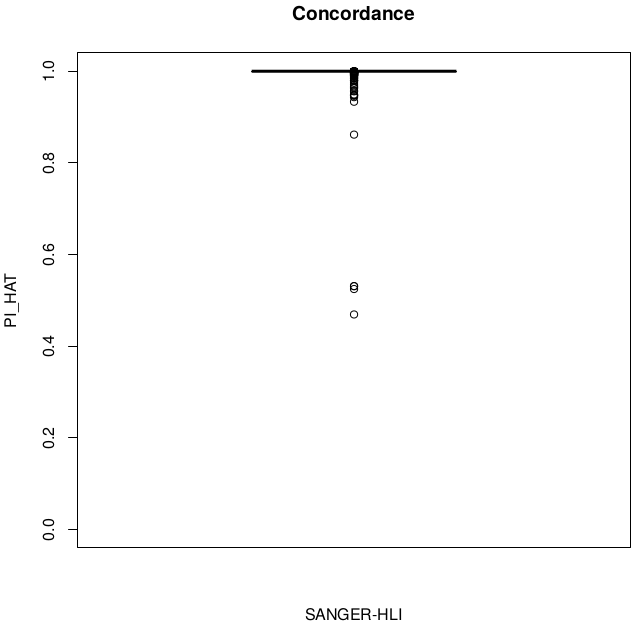
Concordance
SUM(I(PI_HAT<0.9)) = 22
SUM(I(PI_HAT<0.9)) = 5
low het individual
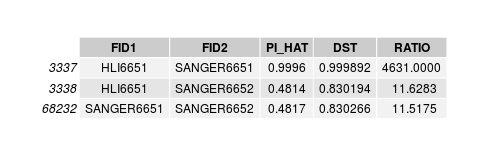
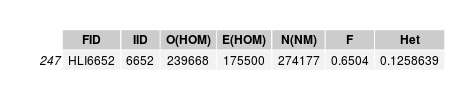
Individual is unrelated to any sample, including its genotyped image
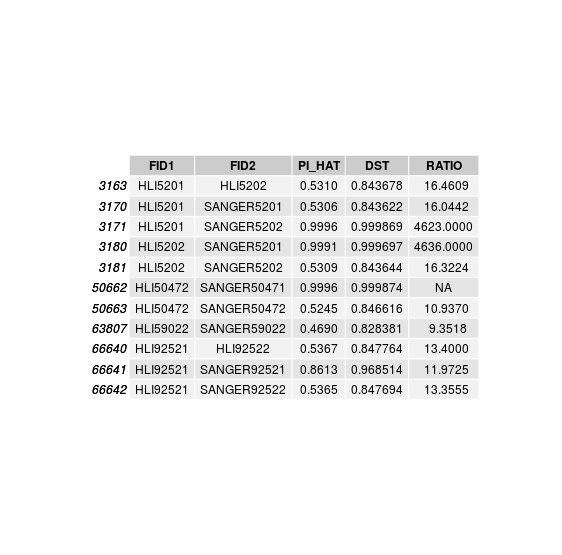
disconcordance analysis
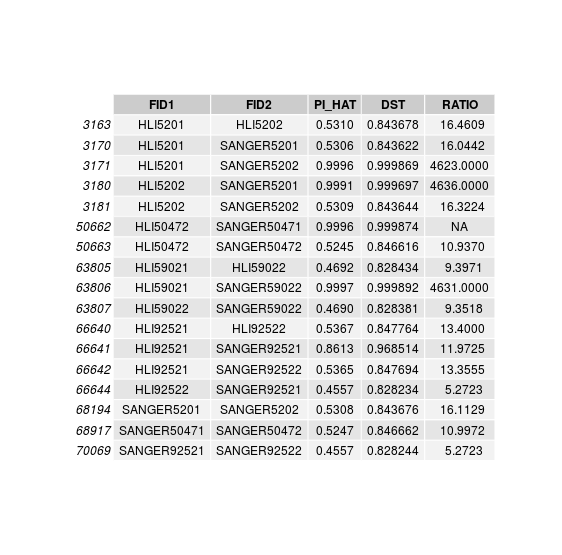
Discordant individuals: 5201, 5202, 50472, 59022, 92521
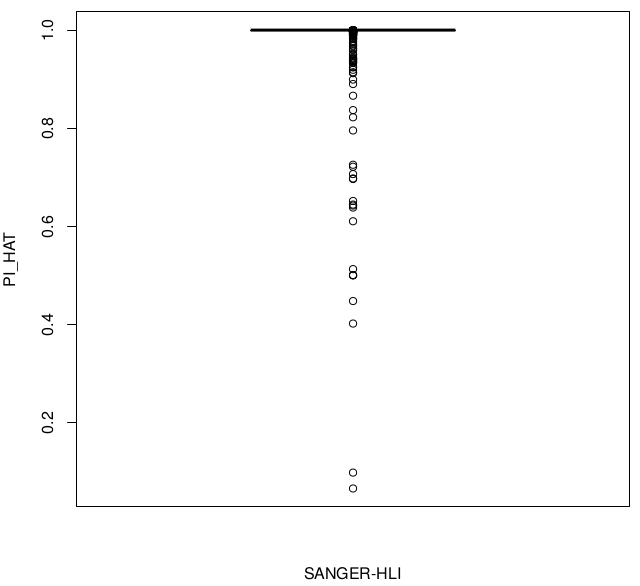
Sample swaps & low concordance
Sample swap within twin pairs:
HLI 5202 is actually SANGER 5201
HLI 5201 is actually SANGER 5202
Sample swaps in unpaired twins:
HLI 50472 is actually SANGER 50471
HLI 59021 is actually SANGER 59022
Individuals with low relatedness with SANGER sample:
HLI 92521 is matched to SANGER 92521 but with lower relatedness.
Unpaired twins either in SANGER of HLI:
SANGER59021
HLI50471
Comparison PLINK vs VCF
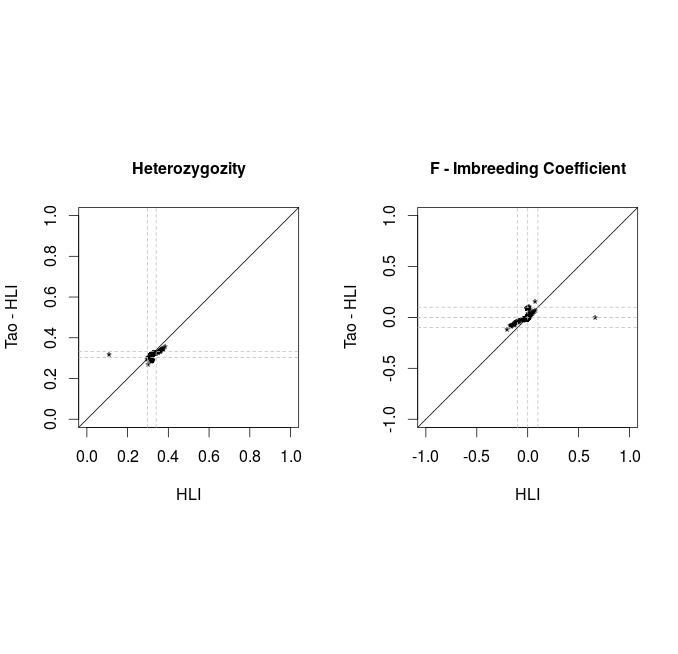
Comparison PLINK vs VCF
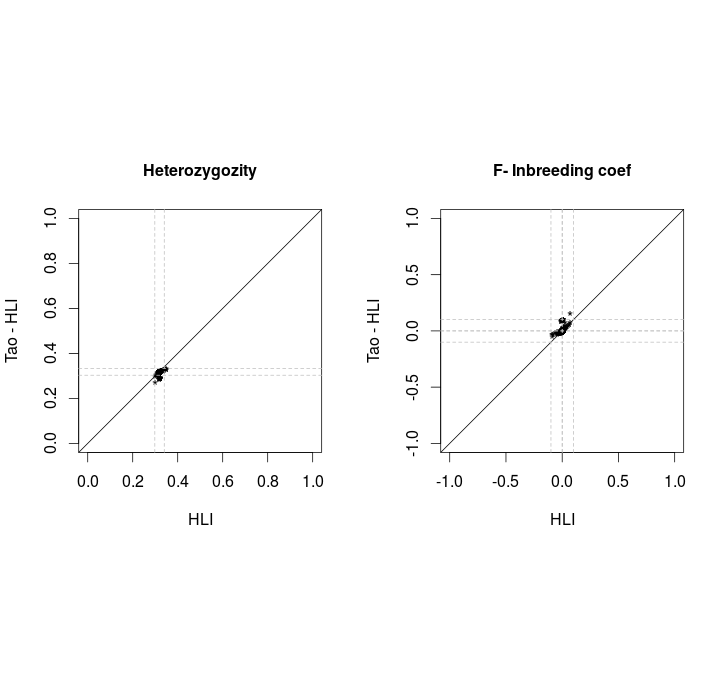
After removal of 16 individuals with |F|>.1
Comparison PLINK vs VCF
Before filtering SNP with MAF<0.05 and HWE<1E-6
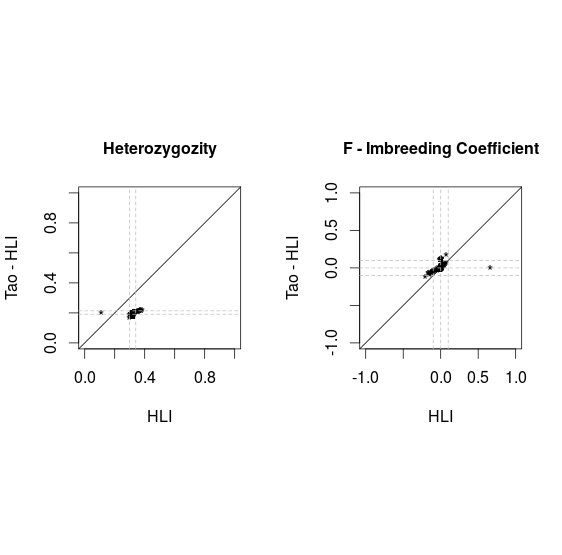
Missing vcf file
[alvesa@athena HLI_HG38]$ ls | grep 6052
60521.vcf.gz
60521.vcf.gz.tbi
Client.Subject.ID Gender Ethnicity Birth.Date FamilyID Relation Zygosity Stool.Sample HLI.genome.ID 1731 60522 F White 10/4/35 762 Twin MZ TRUE 176500025 |
|
|
|