Some deep learning methods
for single-cell NGS data
Sasha Galitsyna
agalitzina@gmail.com
Single-cell NGS methods
- scDNA-Seq (2012): genomic DNA
- scRNA-Seq (2009): transcriptome
- scBS-Seq (2013): methylation
- scATAC-Seq (2015): accessibility
- scDNase-Seq (2015): accessibility
- scChIP-Seq (2015): protein binding
- scNMT-Seq (2018): joint RNA,
- methylation, accessibility
- scHi-C (2013): chromatin interactions
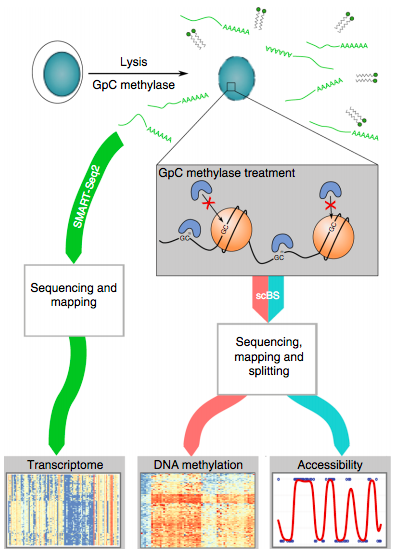
Clark et al. NatCom 2018
Single-cell NGS methods
- scRNA-Seq (2009): transcriptome
- scDNA-Seq (2012): genomic DNA
- scBS-Seq (2013): methylation
- scHi-C (2013): chromatin interactions
- scATAC-Seq (2015): accessibility
- scDNase-Seq (2015): accessibility
- scChIP-Seq (2015): protein binding
- scNMT-Seq (2018): joint RNA+methylation
+accessibility - scCAT (2019): joint accessibility
+transcriptome

Clark et al. NatCom 2018
Single-cell NGS methods
Liu et al. NatCom 2019
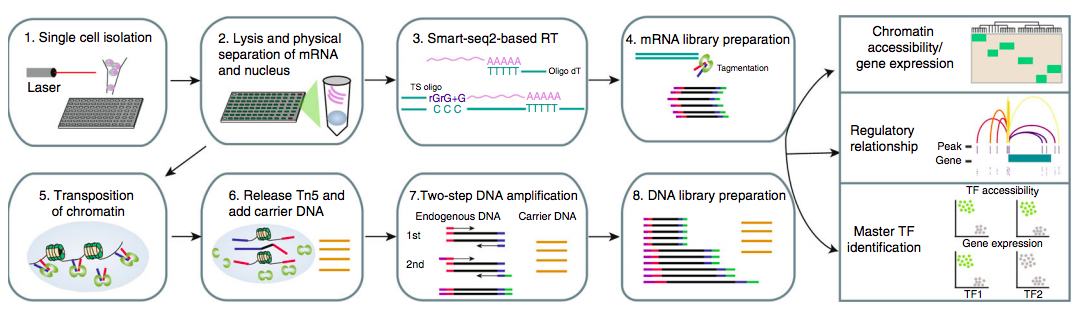
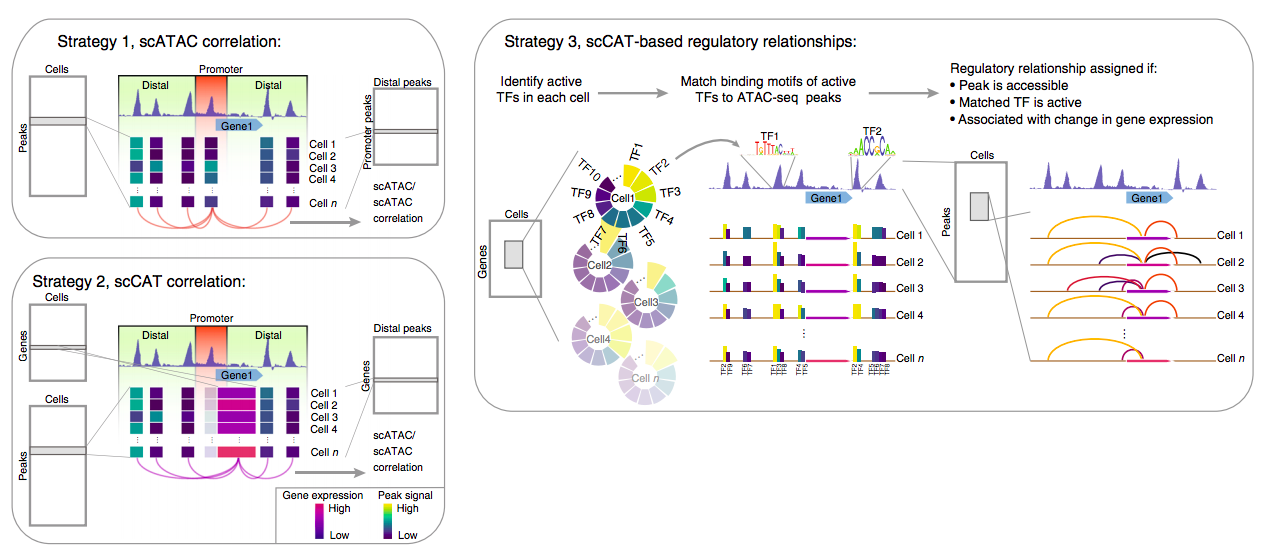
Tasks in sc NGS methods
Hwang et al. 2018
- Data imputation
- Differential expression analysis
- Cell type identification
- Detection of rare cell types
- Cell hierarchy reconstruction
- Inference of regulatory networks
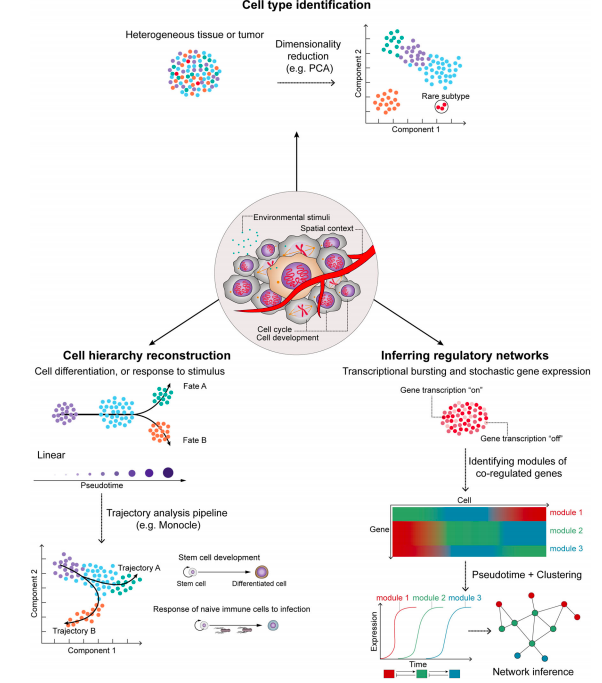
Confounding factors in scNGS
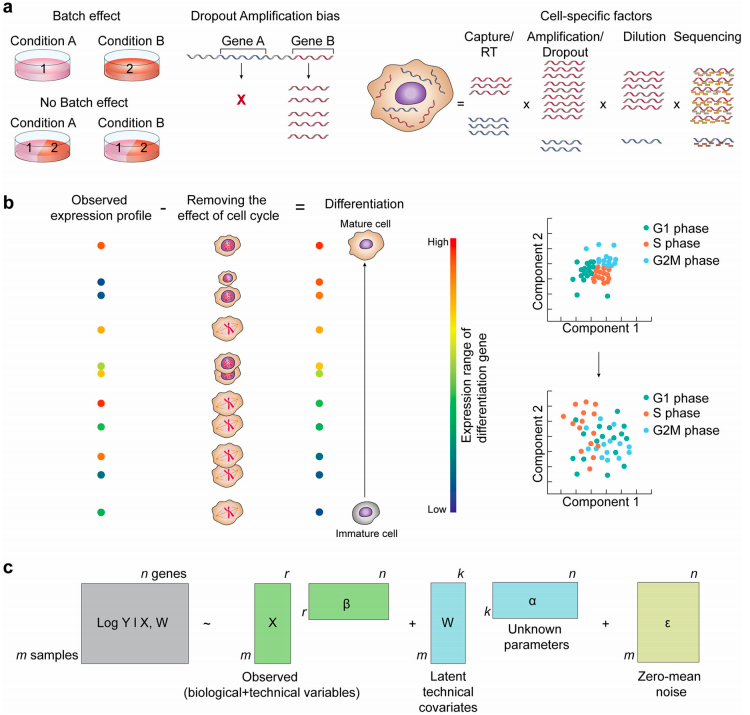
Hwang et al. 2018
Tasks in sc NGS methods

Hwang et al. 2018
- Data imputation
- Differential expression analysis
- Cell type identification
- Detection of rare cell types
- Cell hierarchy reconstruction
- Inference of regulatory networks
Recent advances...
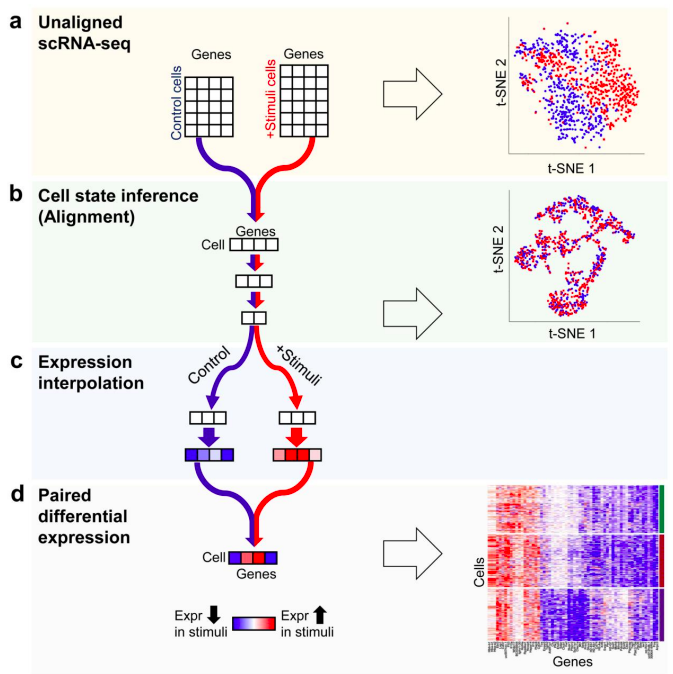
scBLAST (Cao 2019 bioRxiv)
BS-Seq: bisulfite sequencing
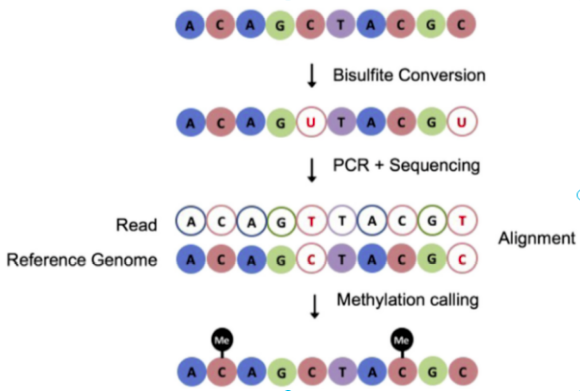
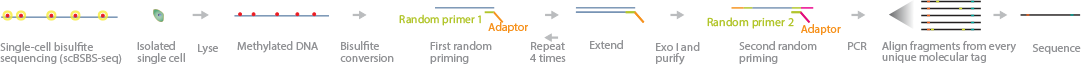
DeepCpG: methylation prediction in single cells
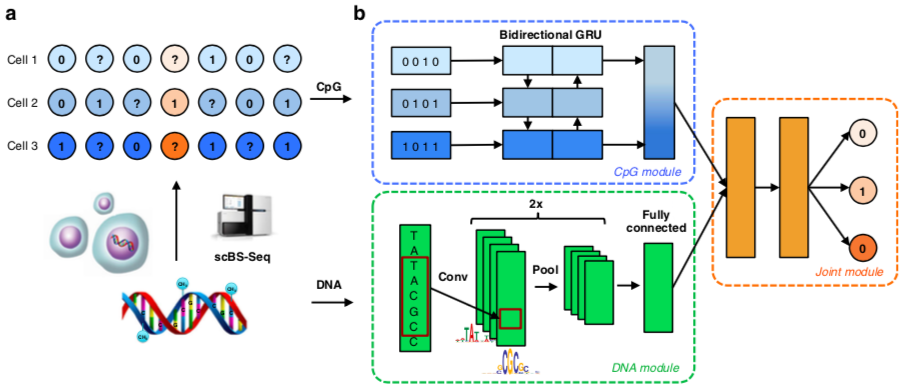
Angermueller et al. 2017 Genome Biology
https://github.com/cangermueller/deepcpg.git
DeepCpG: tasks
Angermueller et al. 2017 Genome Biology

DeepCpG architecture

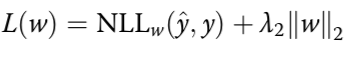
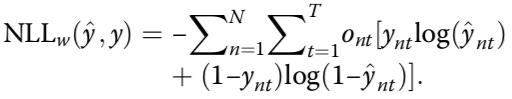
Other methods
- WinAvg
(3001 bp)
- CpGAvg
- Random Forest
(k-mers in 1001 bp window, CpG state in 25neighbors )
- Random Forest from Zhang
(2 CpGneighbors , genomic context, histone modifications, DHS)
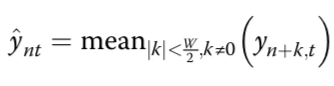
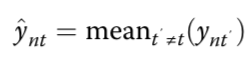
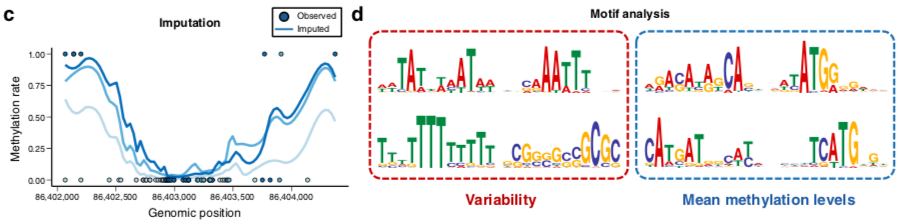
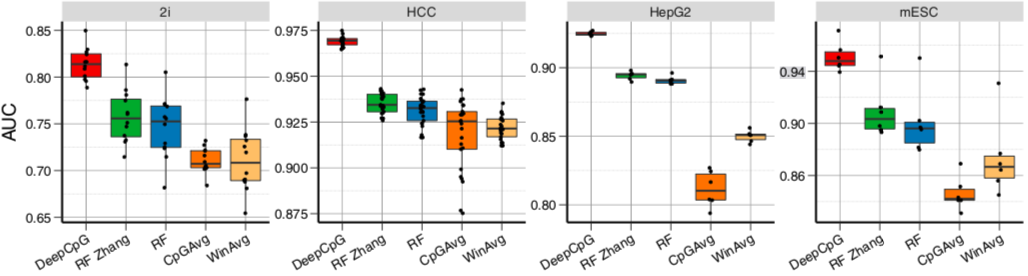
DeepCpG performance
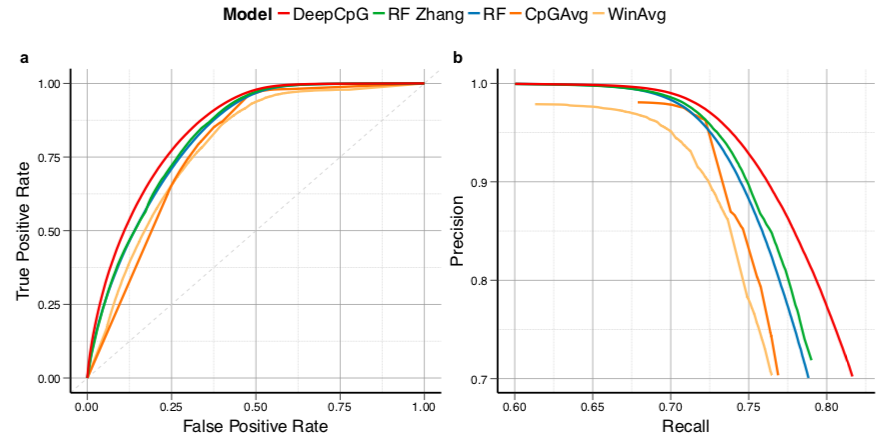
DeepCpG performance
Angermueller et al. 2017 Genome Biology
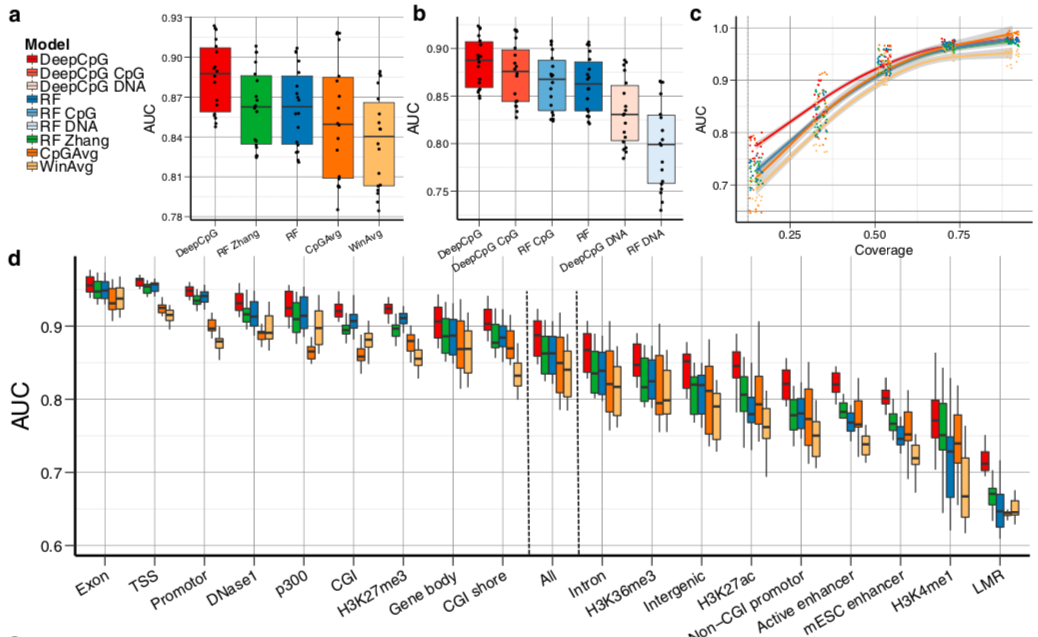
DeepCpG performance
Motifs analysis
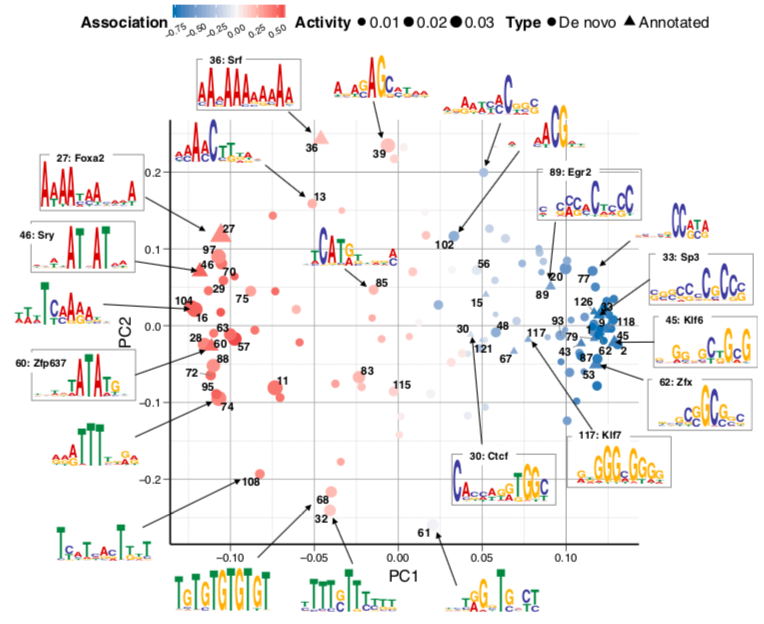
Motifs analysis
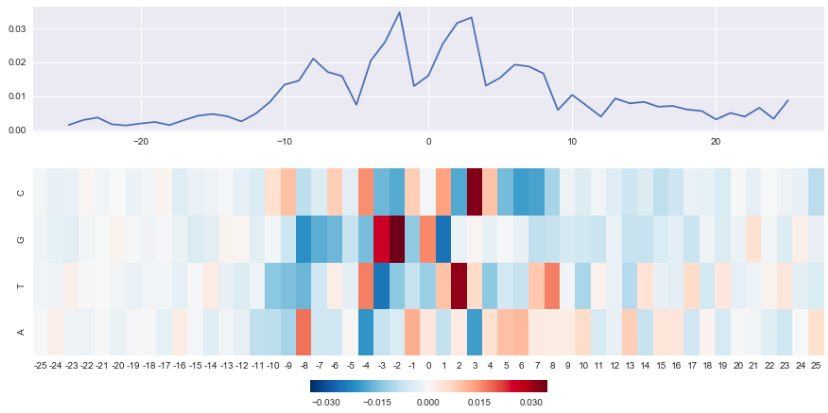
Effect of mutations
Modified model to predict mean and variance
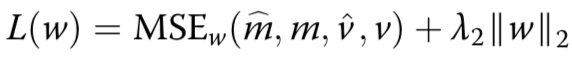
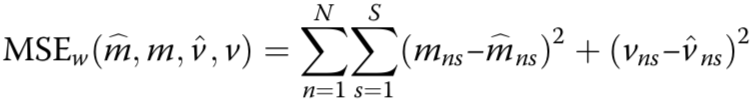
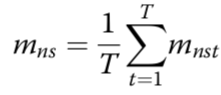
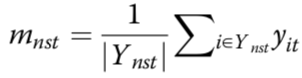
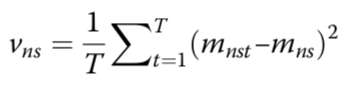
estimated mean methylation rate of cell t computed by averaging the binary methylation state of all observed CpG sites in window s
estimated mean methylation level for a win- dow centred on target site n of a certain size indexed by s
cell-to-cell variance
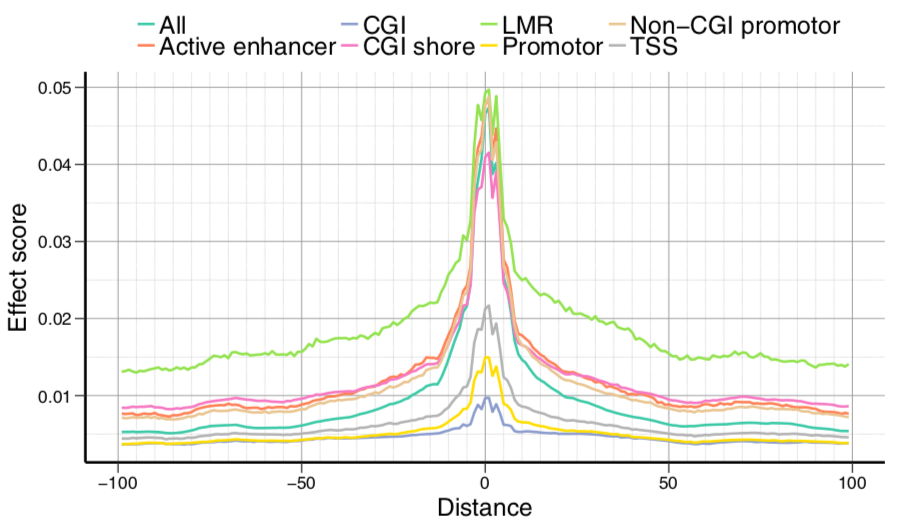
Modified model results
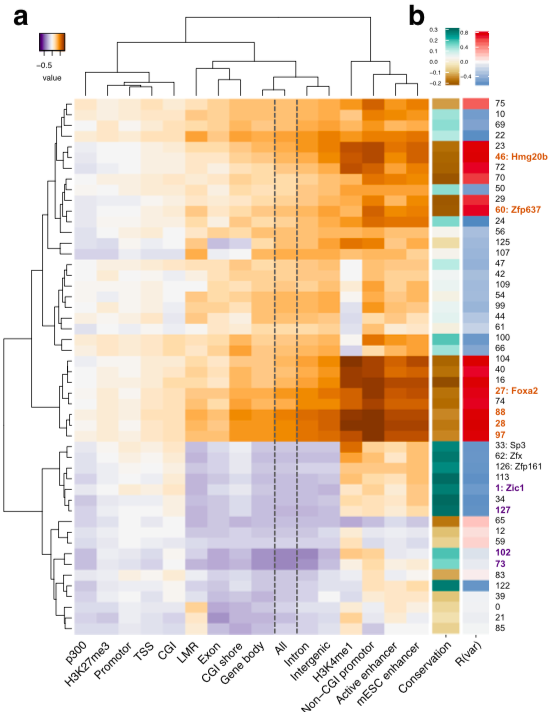
Right: motif effects
brown: association with variance
purple: influence on mean
Left:
correlation between activity vs
conservation or variation
Modified model results
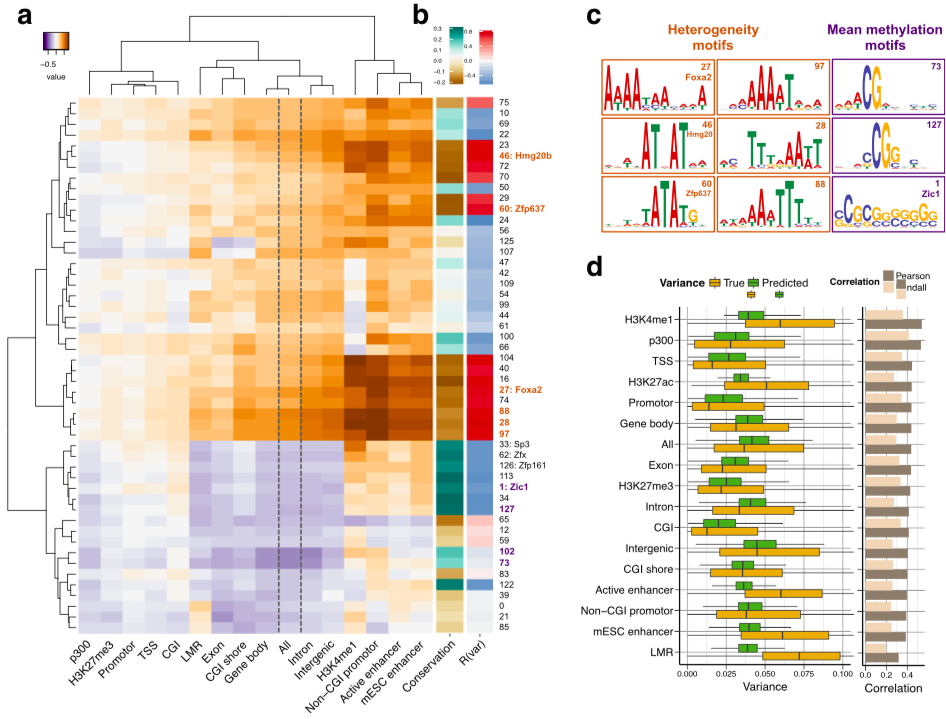
Modified model results

scVI: variational inference of RNA-Seq
Lopez et al. 2018 December Nature Methods
scRNA-Seq data problem:
https://hemberg-lab.github.io/scRNA.seq.course/biological-analysis.html
Svensson bioRxiv 2019
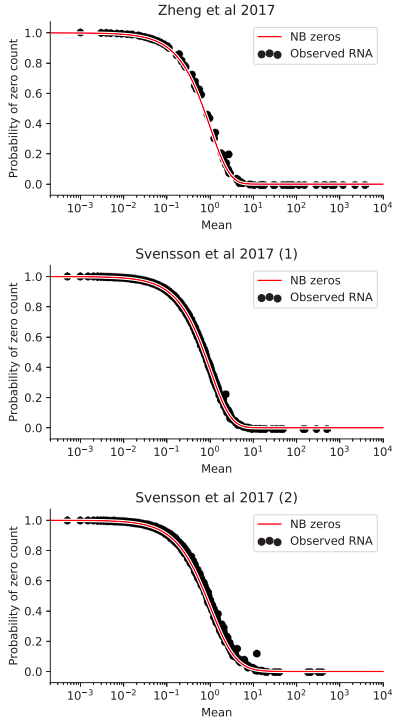
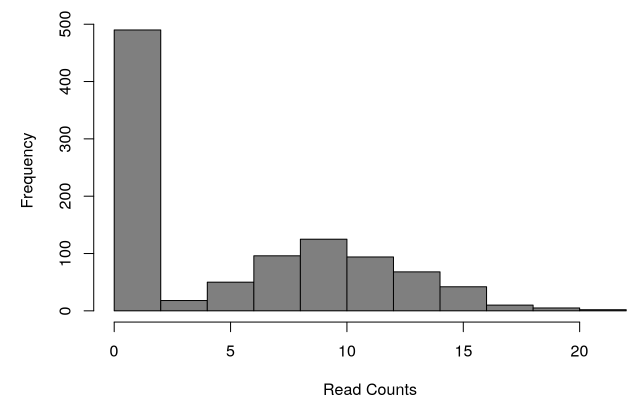
Zero-inflated distribution of counts:
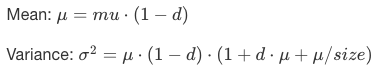
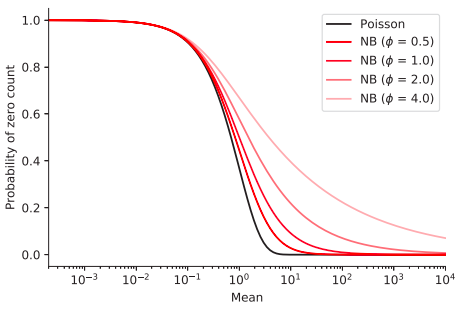
Zero-inflated negative binomial:
Variational autoencoders
for scRNA-Seq embedding
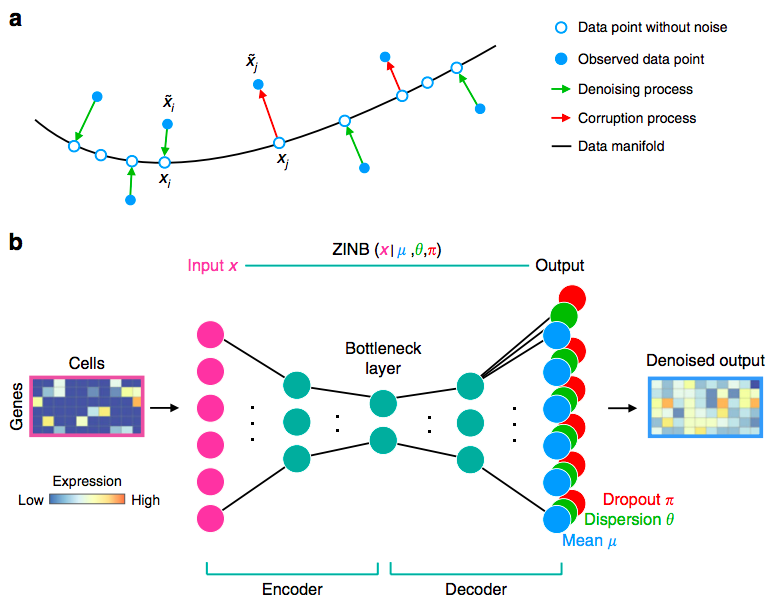
- scvis, scVAE, VASC, DCA: do not distinguish technical and biological effects
- Example for DCA (deep count autoencoder network):
Variational autoencoders
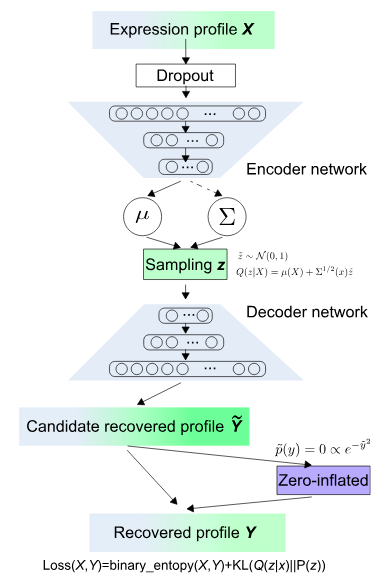
for scRNA-Seq embedding
- Example for VASC:
scVI
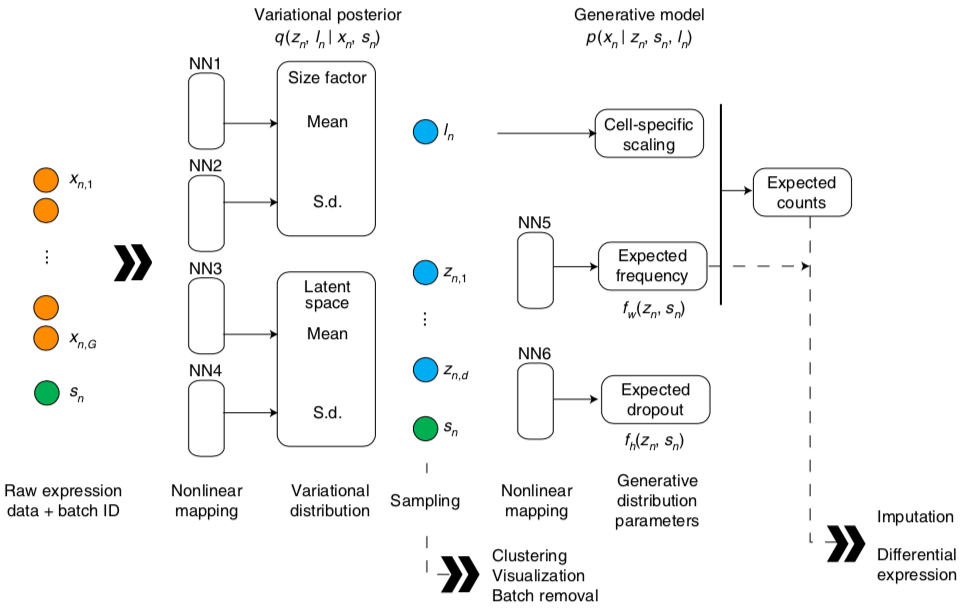

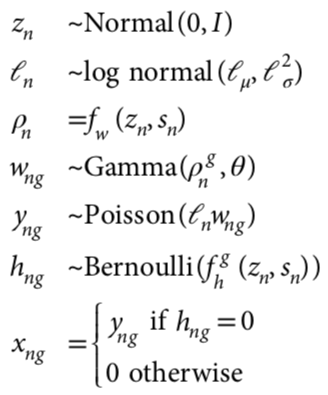
n - cells
g - genes
multivariate normal prior
prior for log-scaling factor for library size
latent variables:
scANVI
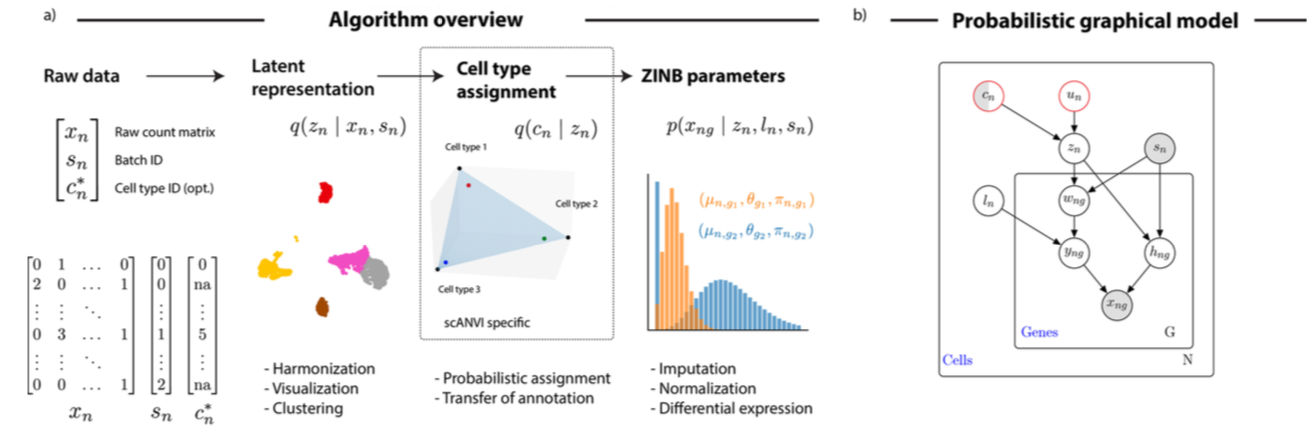
Variables:
Grey: observed
Semi-shaded: ovserved or random
White: latent
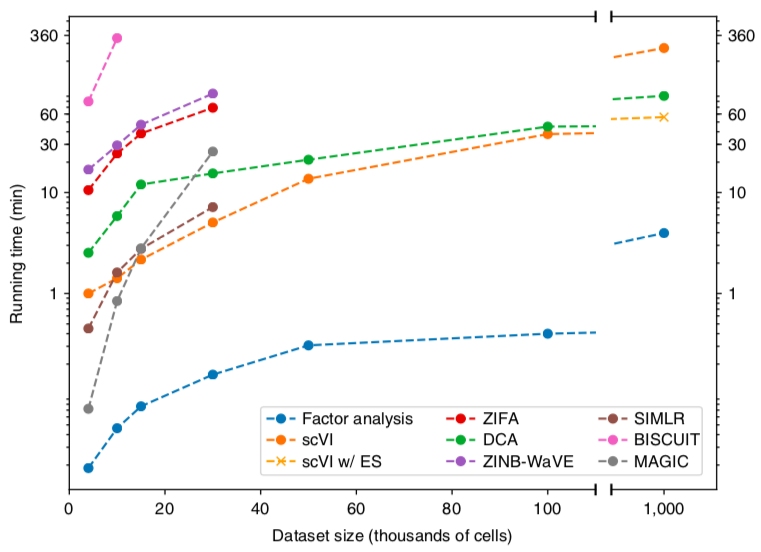
scVI runtime
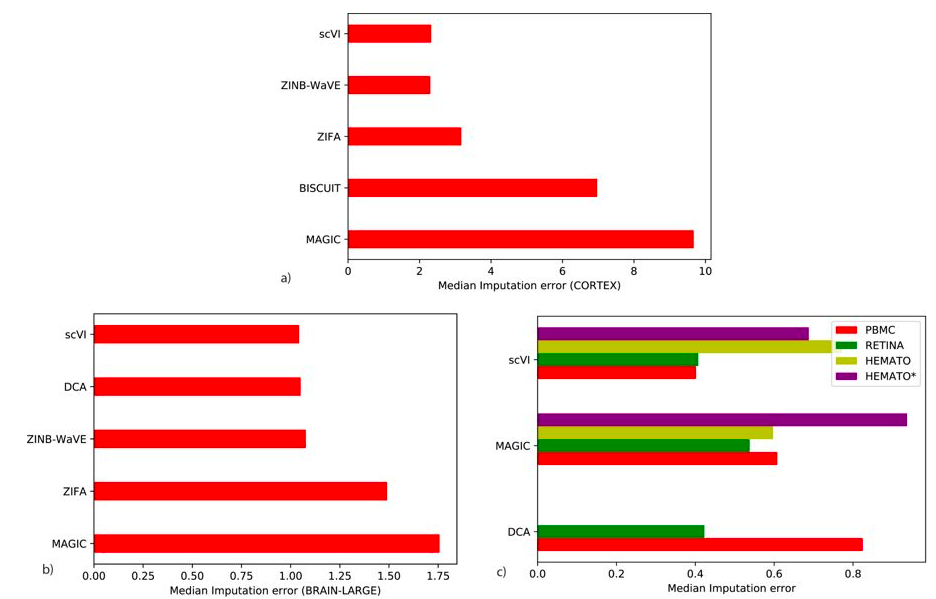
Imputation errors assessment
SIMLR:
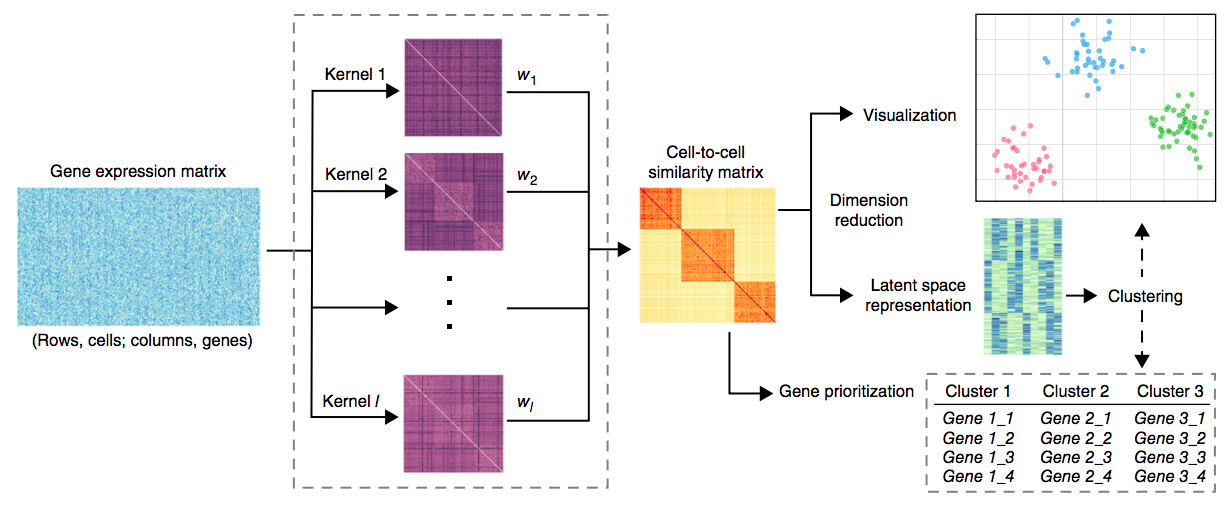
single-cell interpretation via
Wang et al. 2017 Nature Methods
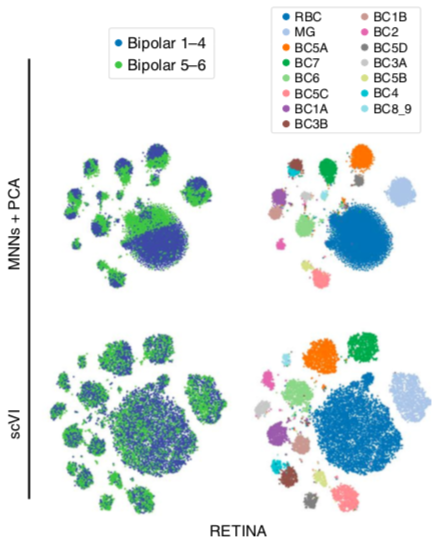
scVI latent space
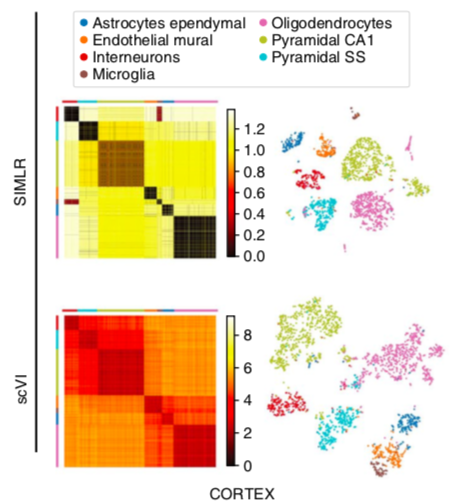
scVI latent space
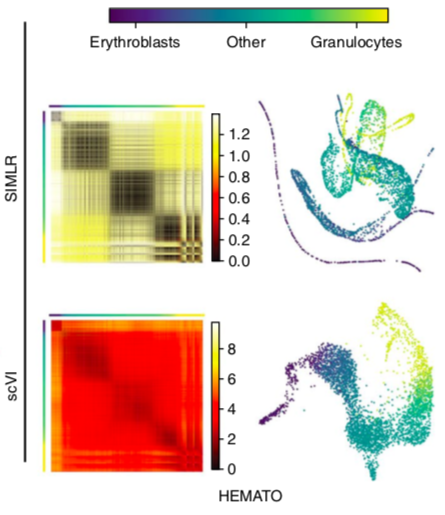
scVI latent space
scVI: differential expression benchmarking
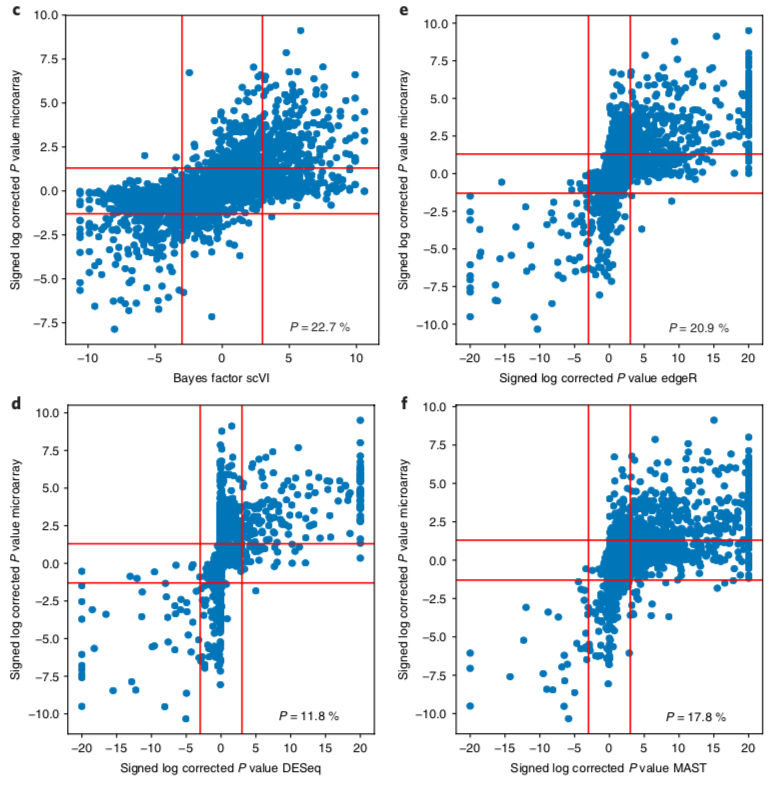
scVI: differential expression benchmarking

scVI: differential expression benchmarking
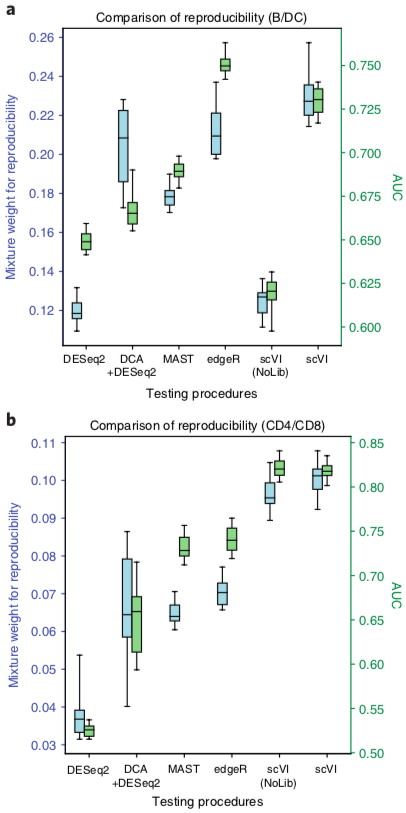
scVI: differential expression benchmarking

See also:
- single-cell ANnotation using Variational Inference (
scANVI ) - Jan 2019, bioRxiv, Xu et al. (the same authors)