KM-ε: A novel Categorical Clustering Algorithm
Mohamad Amin Mohamadi
Prof. Pasi Franti
Summer 2019
Cluster Analysis
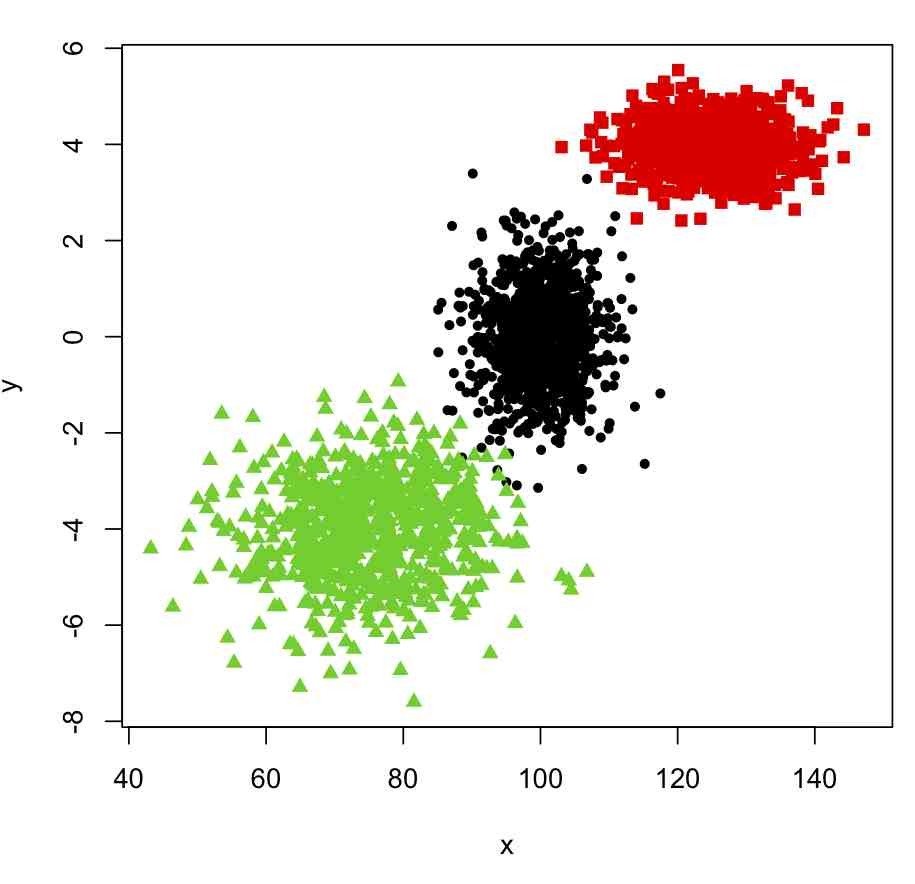
Categorical Data
Related Work
- K-Means like iterative point of view
- Agglomerative point of view
- Direct optimization point of view
- Subspace categorical clustering
- Fuzzy categorical clustering
Motivation
- Lack of optimal convergence guarantee and dependence of the solution to the initialization
- High time complexity for optimal convergence (quadratic for special cases or more for general algorithms)
- Need for extra convergence parameters
- Need for pre or post processing of clustering results
Idea
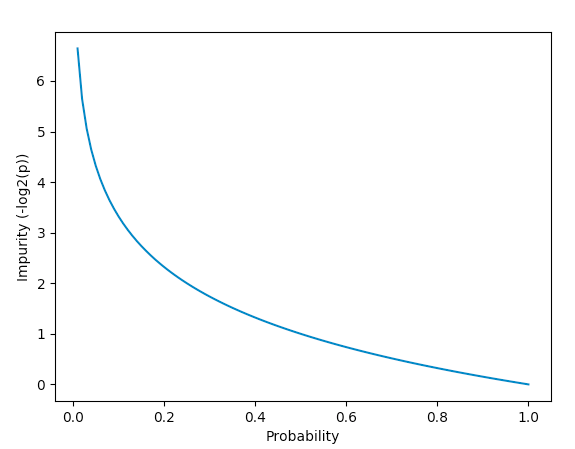
Objective Function
Using entropy as a measure of
purity in clusters
K-means Like Algorithm for Categorical Data
1. Centroid Step:
Calculate probabilities of each category in each attribute for clusters as for each cluster, where is the maximum number of categories in attributes.
2. Assignment step:
Assign each data point to the cluster where is:
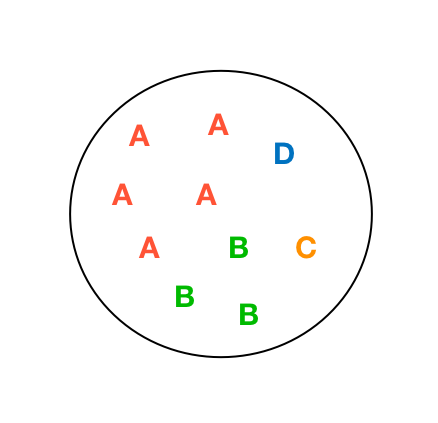
A Toy Example
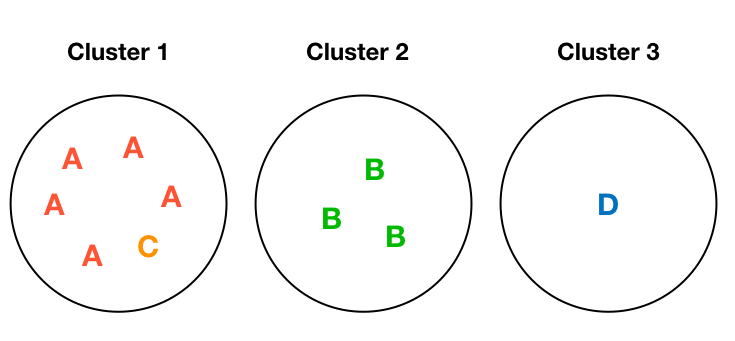
Entropy: 0
Entropy: 0
Entropy: 0.45
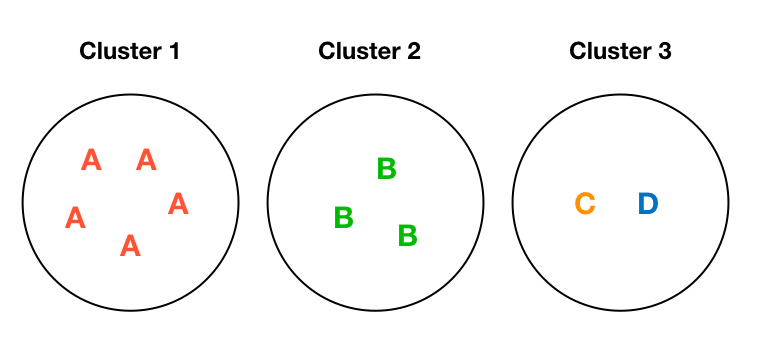
A Toy Example (Optimum)
Entropy: 0
Entropy: 0.69
Entropy: 0
Calculating Centroids
| P(A) | P(B) | P(C) | P(D) | |
|---|---|---|---|---|
| Cluster 1 | 0.83 | - | 0.17 | - |
| Cluster 2 | - | 1.00 | - | - |
| Cluster 3 | - | - | - | 1.00 |

Assigning Datapoints
| Data | Cluster 1 | Cluster 2 | Cluster 3 | Selected |
|---|---|---|---|---|
| 0.18 |
|
1 | ||
| 0 | 2 | |||
| 1.79 | 3 | |||
| 0 | 1 |
We select the best cluster for each datapoint according to equation (1).
Assigning Datapoints
Problem:
Log of 0 is infinite, so the datapoints won't move if D-1 attributes feel comfortable about a move, but only one attribute doesn't!
Motivation:
Decrease the influence of categories with zero probability. Anticipate newcomers in less crowded clusters!
No datapoint moves! Each datapoint has it's best situation (according to eq. 1) in its own cluster!
Epsilon Strategy
A
A
A
A
A
A
B
B
B
B
B
B
B
D
E
E
F
C
?
?
?
B
A
* Costs Calculated according to equation 1
Epsilon Strategy
Calculating Centroids (Epsilon Strategy)
Let's start with a simple epsilon:
Calculating Centroids (Epsilon Strategy)
| P(A) | P(B) | P(C) | P(D) | |
|---|---|---|---|---|
| Cluster 1 | 0.67 | 0.17 | 0.17 | 0.17 |
| Cluster 2 | 0.25 | 0.75 | 0.25 | 0.25 |
| Cluster 3 | 0.5 | 0.5 | 0.5 | 0.5 |

Assigning Datapoints (Epsilon Strategy)
| Data | Cluster 1 | Cluster 2 | Cluster 3 | Selected |
|---|---|---|---|---|
| 0.40 | 1.38 | 0.69 | 1 | |
| 1.77 | 0.28 | 0.69 | 2 | |
| 1.77 | 1.38 | 0.69 | 3 | |
| 1.77 | 1.38 | 0.69 | 3 |
We select the best cluster for each datapoint according to equation (1).
Assigning data points with epsilon idea
- Datapoint C moves from cluster 1 to cluster 3
- No other datapoints move afterwards
- Reached the global optimum!

How To Choose Epsilon?
| Strategy | Average Impurity | Impurity SE |
|---|---|---|
| 7.59* | 0.31* | |
| 7.49* | 0.26* |
* Over 100 tests on the test dataset
Pseudo-Code: KM-ε
Input
-------------
X (N × D)
k
update_proportion_criterion
Algorithm
-------------
assign datapoints randomly to k clusters
do
centroids = calculate_centroids()
movements = assign_datapoints(centroids)
while movements > update_proportion_criterion * NPseudo-Code: KM-ε
function calculate_centroids()
for j in 1..k
cluster_data = items in cluster j // n_j * D matrix
for d in 1..D
if there is empty category in dimension d of cluster_data:
for each category in dimension d as cat:
if count(cat) in cluster_data > 0:
centroids[k][d][cat] = count(cat) / (n_j + 1)
else:
centroids[k][d][cat] = 1 / (n_j + 1)
else:
for each category in dimension d as cat:
centroids[k][d][cat] = count(cat) / n_j
return centroidsPseudo-Code: KM-ε
function assign_datapoints(centroids)
movements = 0
for i in 1..N:
selected_cluster = -1
most_entropy = -infinite
for j in 1..k:
cluster_entropy = 0
for d in 1..D:
category = X[i][d]
cluster_entropy -= log(centroids[j][d][category])
if cluster_entropy > most_entropy:
most_entropy = cluster_entropy
selected_cluster = j
if selected_cluster != prev_cluster[i]:
movements++
move datapoint to selected_cluster
return movementsTesting The Algorithm
Mushroom dataset
For testing the algorithm, we used mushroom dataset.
Mushroom dataset properties:
- 8124 rows
- 22 attributes (all categorical)
We used 16 clusters for the mushroom dataset.
Results of running the algorithm on the dataset are available in the next slides.
Empirical results: KM-ε
Results of 100 runs:
| Statistic | Value |
|---|---|
| Average impurity | 7.49 |
| Best (lowest) impurity | 7.04 |
| Worst (highest) impurity | 8.12 |
| Standard error of impurity | 0.26 |
| Empty cluster situations* | 9 |
* By empty cluster situation, we mean that in some point the algorithm created some empty cluster and we couldn't continue the algorithm from there and thus, that case was dumped from the statistics
Visualization: KM-ε
Impurity: 7.41
* Visualization in 2D using TSNE
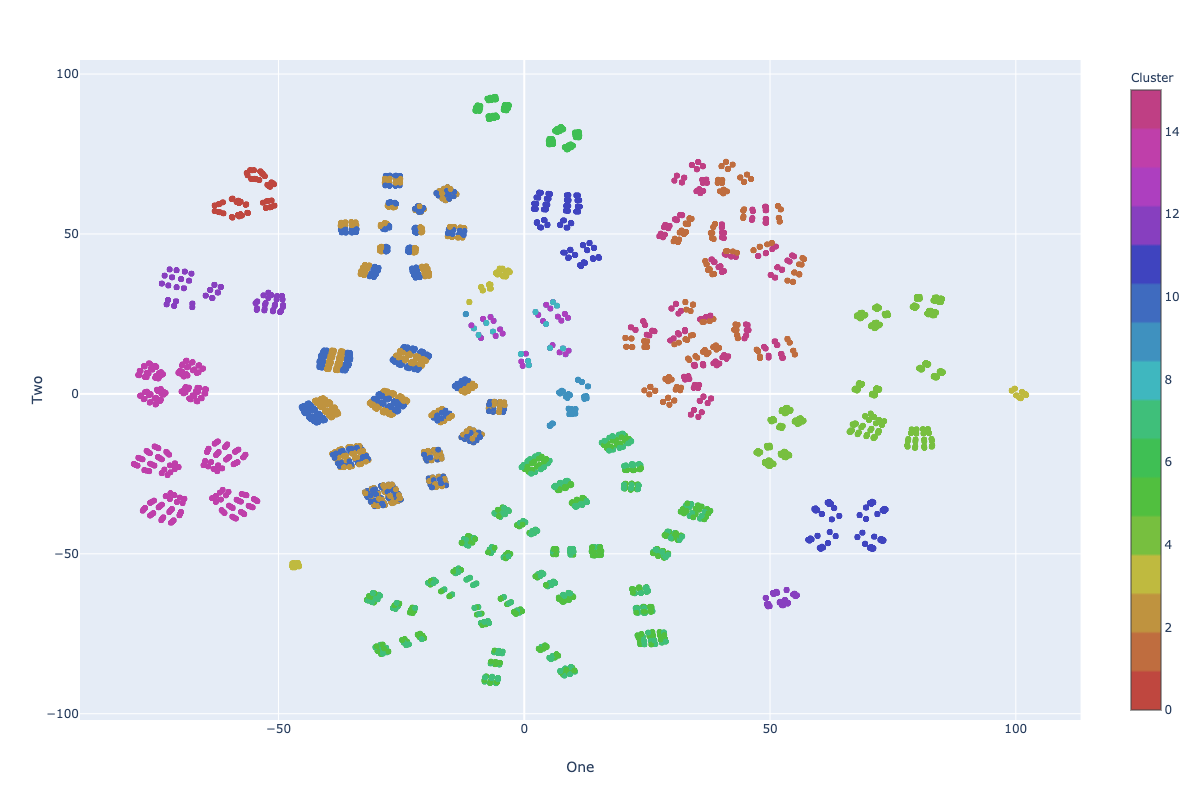
Far From Optimum
Improvement?
Add Random Swap
Random Swap
KM-ε + Random Swap
Result of 100 runs:
| Statistic | Value |
|---|---|
| Average impurity | 7.13 |
| Best (lowest) impurity | 6.98 |
| Worst (highest) impurity | 7.90 |
| Standard error of impurity | 0.13 |
| Empty cluster situations* | 4 |
* By empty cluster situation, we mean that in some point the algorithm created some empty cluster and we couldn't continue the algorithm from there and thus, that case was dumped from the statistics
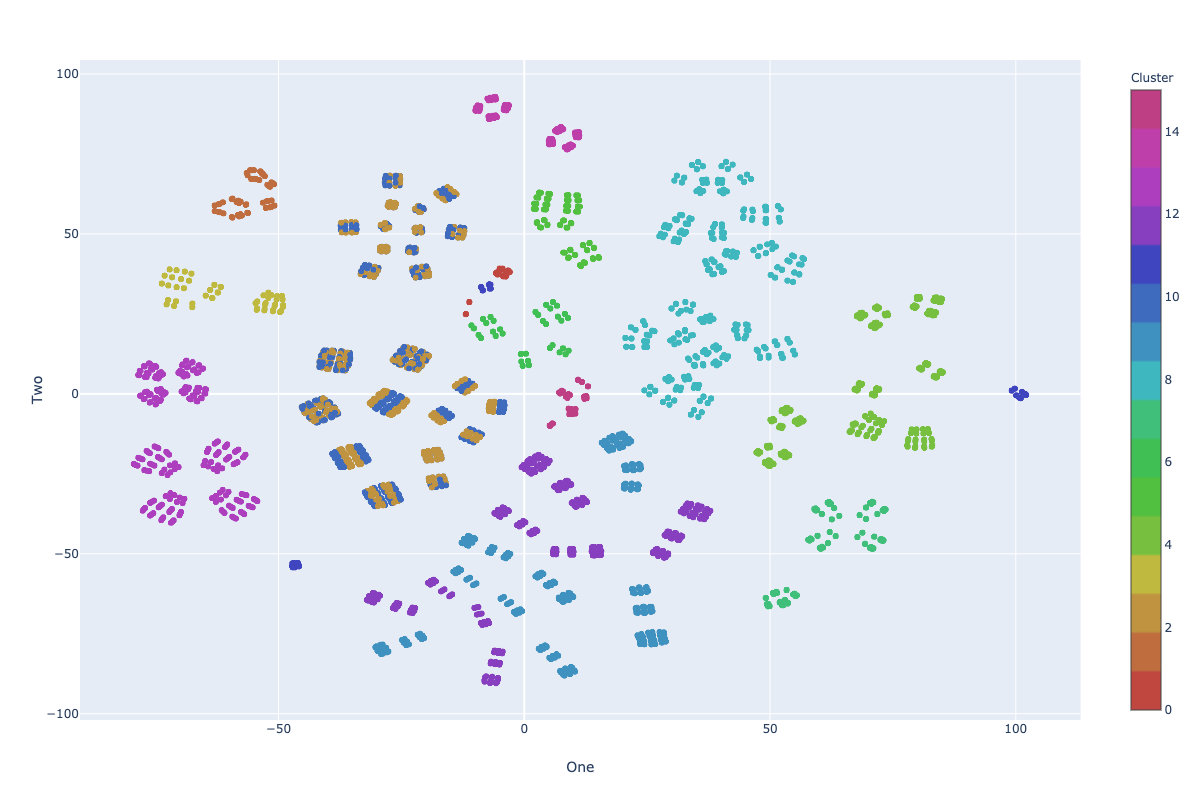
Visualization: KM-ε + Random Swap
Impurity: 7.10
Stress Test: KM-ε + Random Swap
- For checking the rate of convergence of the algorithm, we tested the algorithm with 5000 random swaps to see if it achieves the overall impurity of <= 6.98 (reached with 100 random swap at least once) or not.
- The result was NO!, the impurity achieved using 5000 random swaps was 6.99.
Other Assignment Strategies
- The other assignment strategies tested were:
-
SE Strategy: Assigning the datapoints to the cluster with minimum distance where distance is defined as:
- SE-Full Strategy: Assigning the datapoints to the cluster with minimum distance where distance is defined as
-
SE Strategy: Assigning the datapoints to the cluster with minimum distance where distance is defined as:
* where cats represents the categories in an attribute
Results: SE Strategy
Here are the statistics of algorithm runs on mushroom dataset using the SE strategy assignment .
* By empty cluster situation, we mean that in some point the algorithm created some empty cluster and we couldn't continue the algorithm from there and thus, that case was dumped from the statistics
| K-Means (100 Runs) | K-Means + 100 Random Swaps (50 Runs) | |
|---|---|---|
| Average impurity | 7.51 | 6.99 |
| Best (lowest) impurity | 7.00 | 6.95 (New best) |
| Worst (highest) impurity | 8.79 | 7.07 |
| Standard error of impurity | 0.13 | 0.02 |
| Empty cluster situations* | 27 | 2 |
Stress test visualization: SE Strategy
Impurity: 6.95
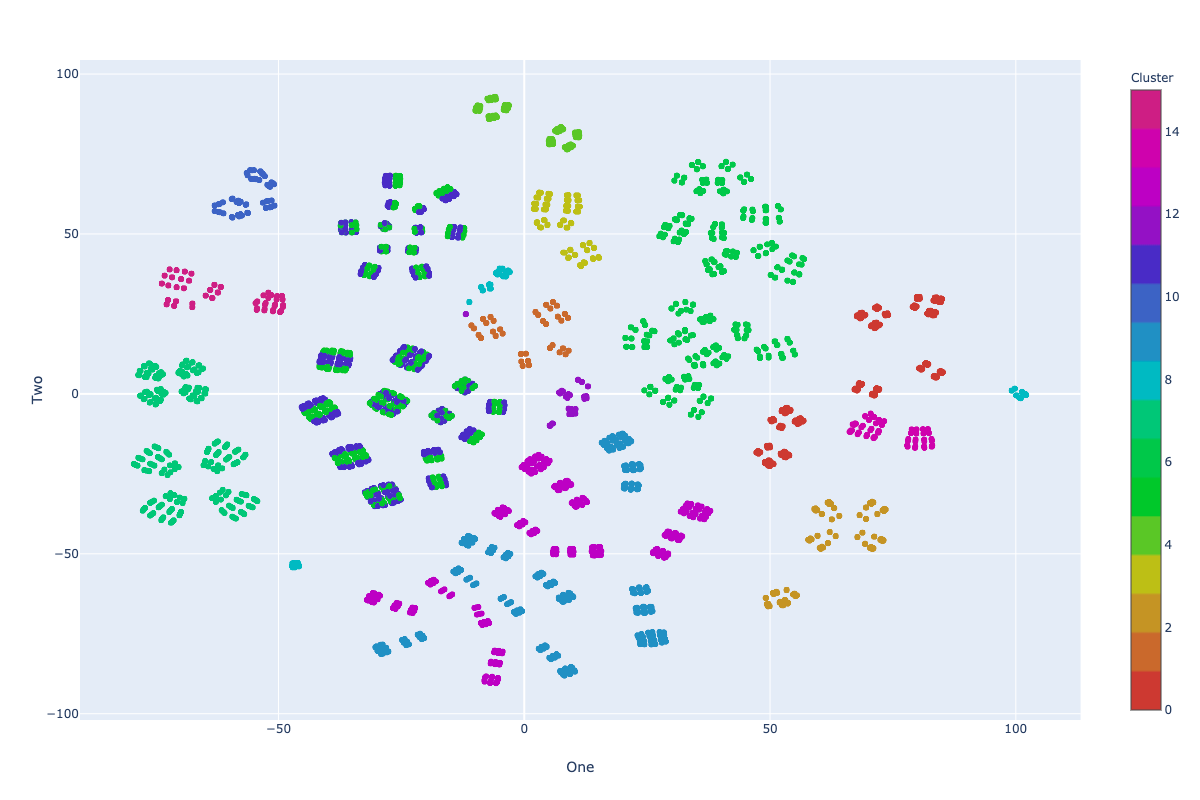
Stress test: SE Strategy
- We reached the overall impurity of 6.95 during our tests with 100 random swaps in this strategy, which is a new best achieved clustering.
- For checking the rate of convergence of the algorithm, we tested the algorithm with 10000 random swaps to see if it achieves the overall impurity of <= 6.95 (reached with 100 random swap at least once) or not.
- The result was Yes!, the impurity achieved using 5000 random swaps was 6.95, which can indicate the stable convergence in SE strategy.
Strategy Comparison
| Impurity | Epsilon = 1/N | SE Strategy |
|---|---|---|
| Average | 7.013 | 6.99 |
| Best (lowest) | 6.98 | 6.95 |
| Worst (highest) | 7.90 | 7.07 |
| Standard error | 0.13 | 0.02 |
Strategy Comparison
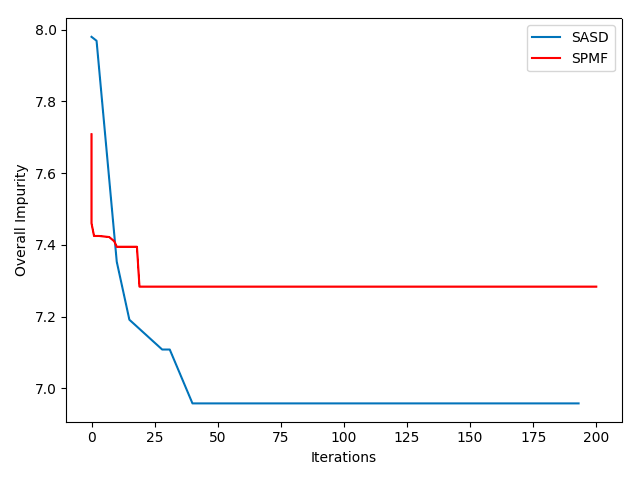
Convergence comparison plot for SE strategy (SASD) and initial epsilon 1/N
Strategy Comparison
According to the statistics achieved using the experiments, it seems like SE strategy has much faster convergence rate compared to the epsilon strategy, and this is probably because of the epsilon not being a good anticipation of the future changes in the next iterations of the algorithm. But in general, a good epsilon should give the best convergence rate.
Internal Evaluation
|
Algorithm |
Soybean M = 4 Cost std |
Mushroom M = 16 Cost std |
SPECT M = 8 Cost std |
|---|---|---|---|
| ACE | 7.83 0 | 7.01 n/a | 8.43 0.07 |
| ROCK | 9.20 0 | n/a n/a | 10.82 0 |
| K-medoids | 10.94 1.57 | 13.46 0.71 | 10.06 0.44 |
| K-modes | 8.66 1.06 | 10.19 1.45 | 9.25 0.29 |
| K-distributions | 9.00 0.93 | 7.87 0.44 | 8.25 0.13 |
| K-represantives | 8.30 0.83 | 7.51 0.30 | 8.64 0.13 |
| K-histograms | 8.04 0.53 | 7.31 0.18 | 8.64 0.15 |
| K-entropy | 8.15 0.56 | 7.31 0.18 | 8.06 0.07 |
| Proposed | 7.83 0 | 6.95 0 | 7.97 0 |
External Evaluations
To be added soon
Conclusion
-
Proposed new algorithm: KM-ε
- Optimal convergence guaranteed with adequate number of random swaps independent of the initialization
- Linear time complexity
- No extra convergence parameters
- No pre or post processing of clusters
-
Future Work
- Better anticipation of ε to have much higher rate of convergence
- Clustering mixed (numeric + categorical) data using the proposed concept