Rhythms of the Hippocampal Network
Laura Lee Colgin, Nature Reviews 2016
Plus a lil bit of REL
& More
The Hippocampal Structures

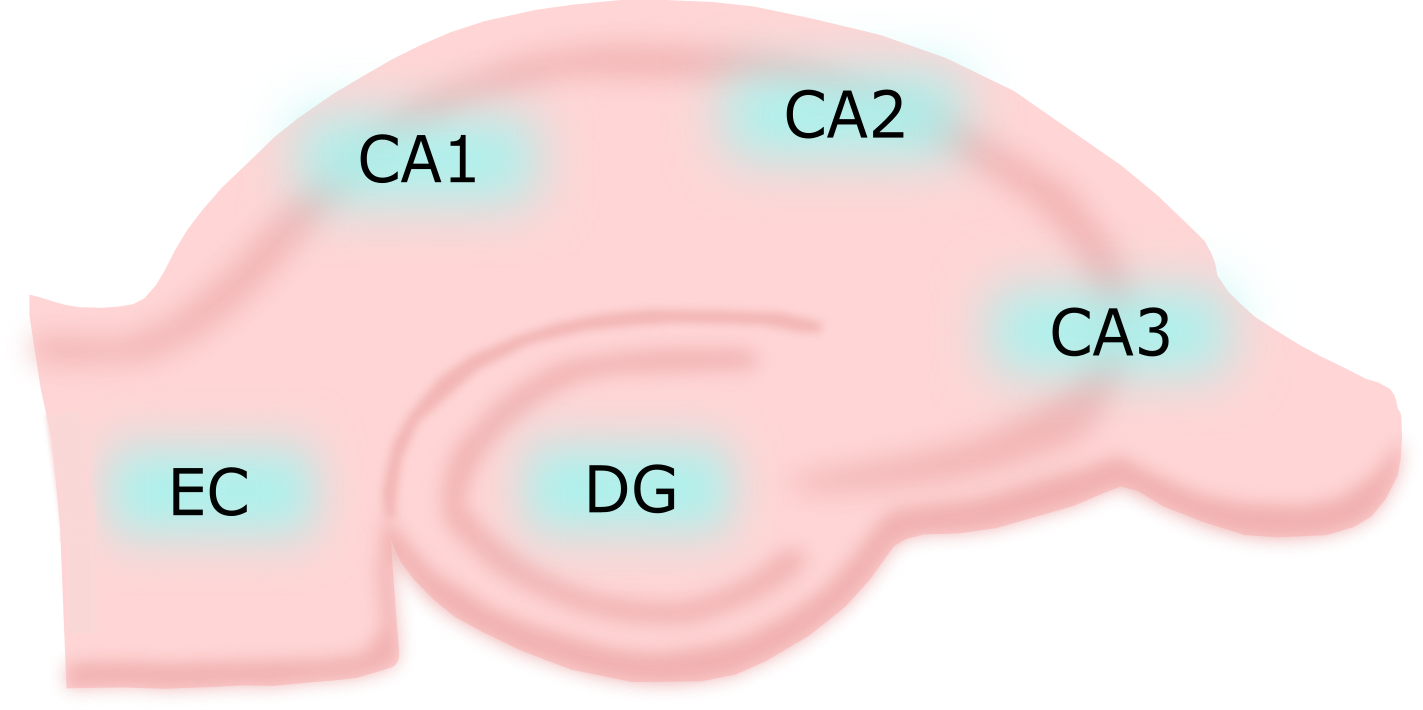
The Hippocampal Neurons

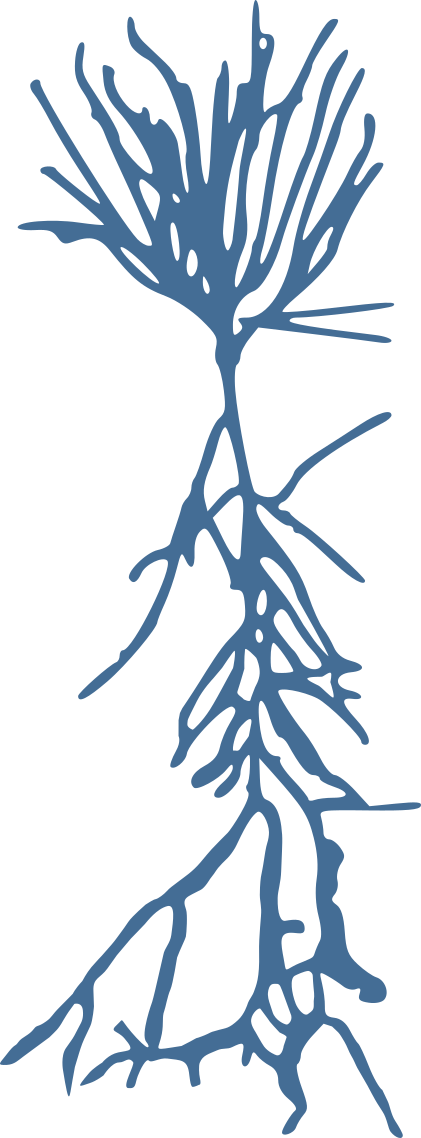
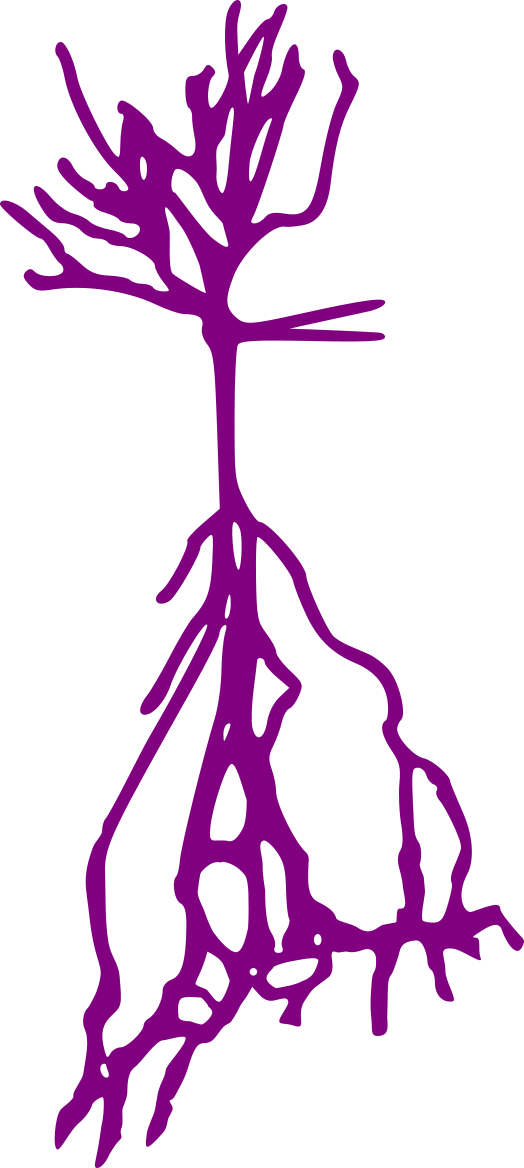
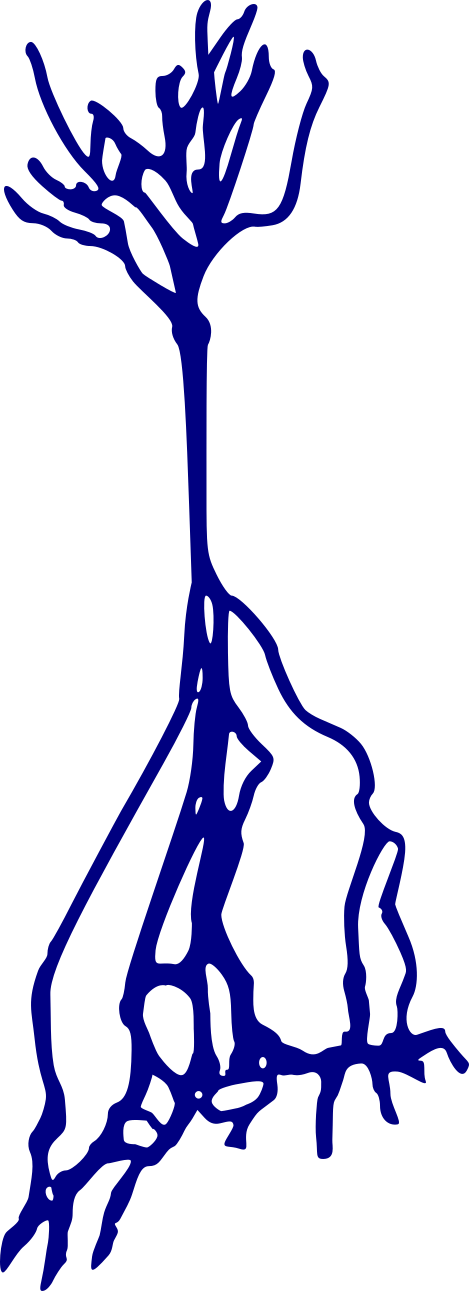

Stellate neurons
Pyramidal neurons

Granule neurons

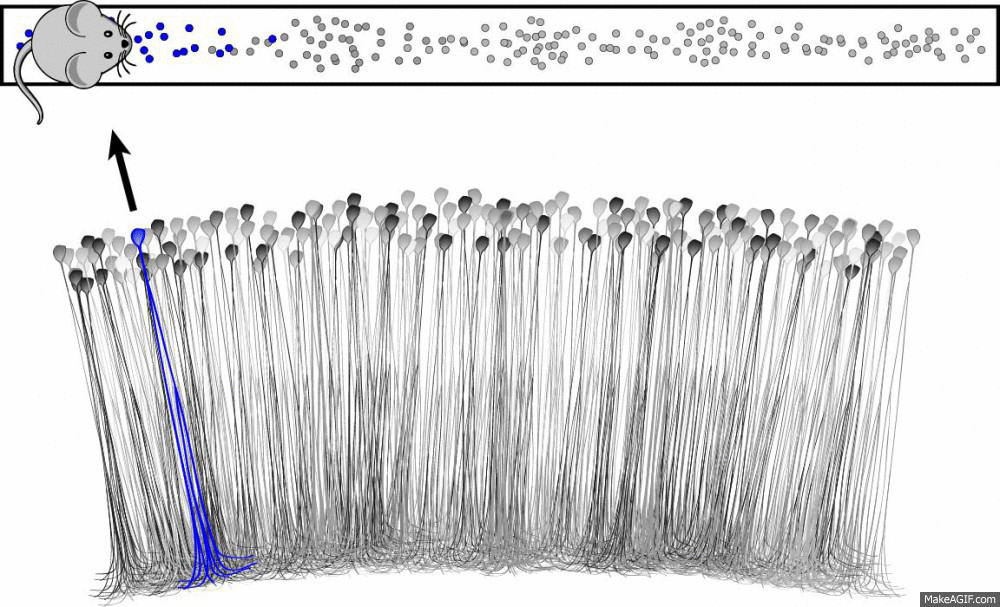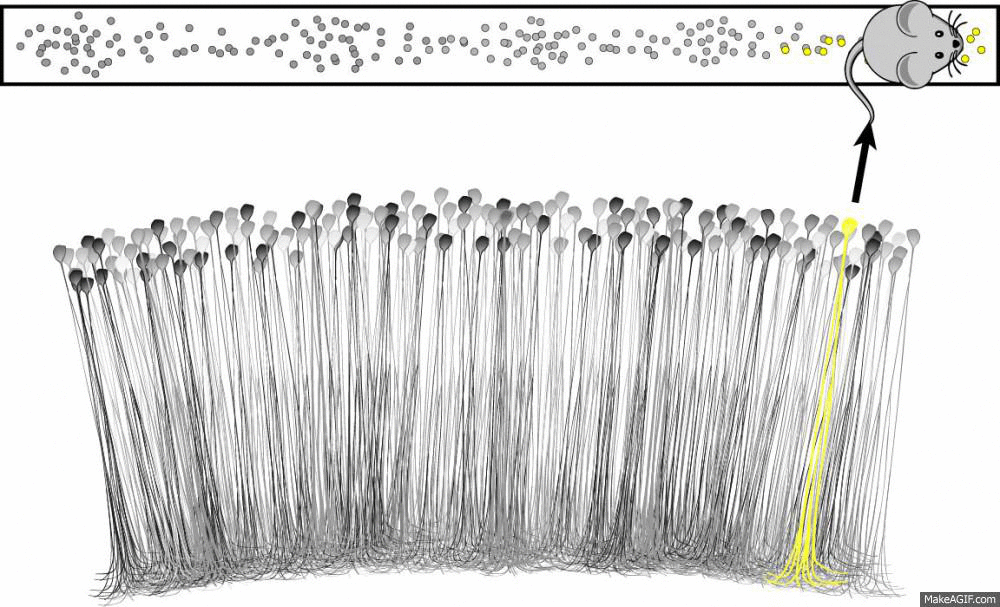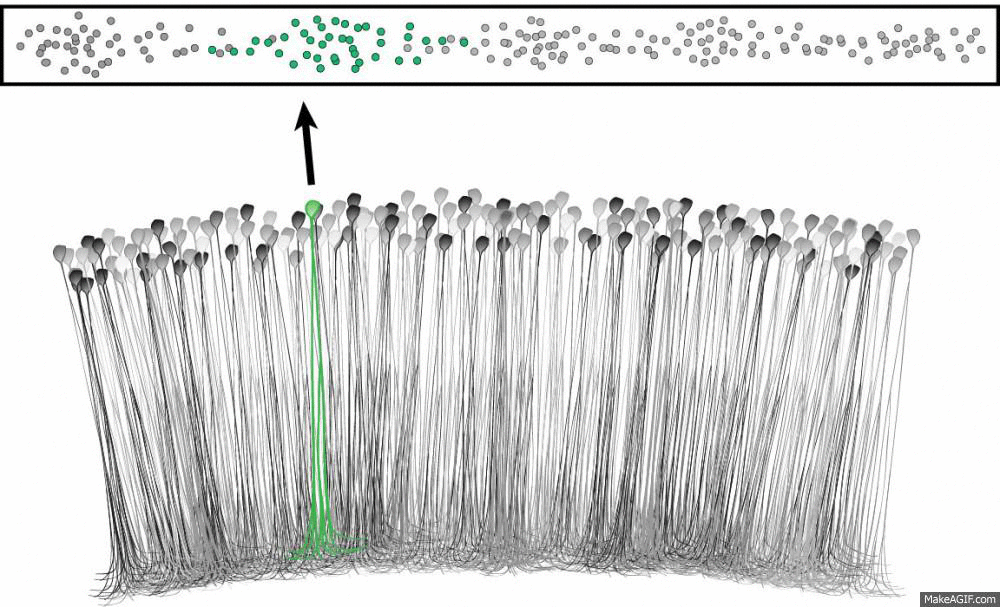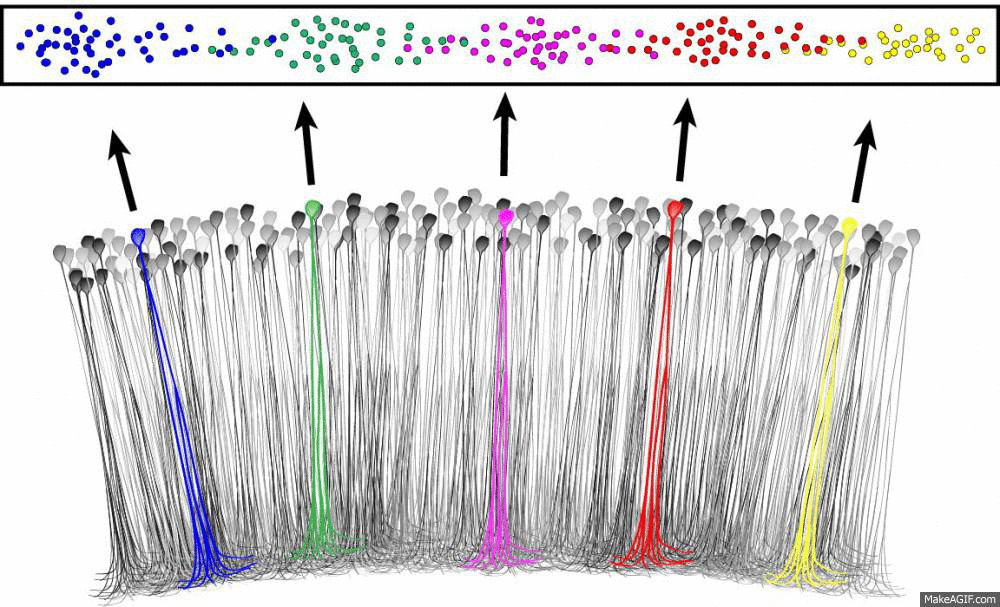
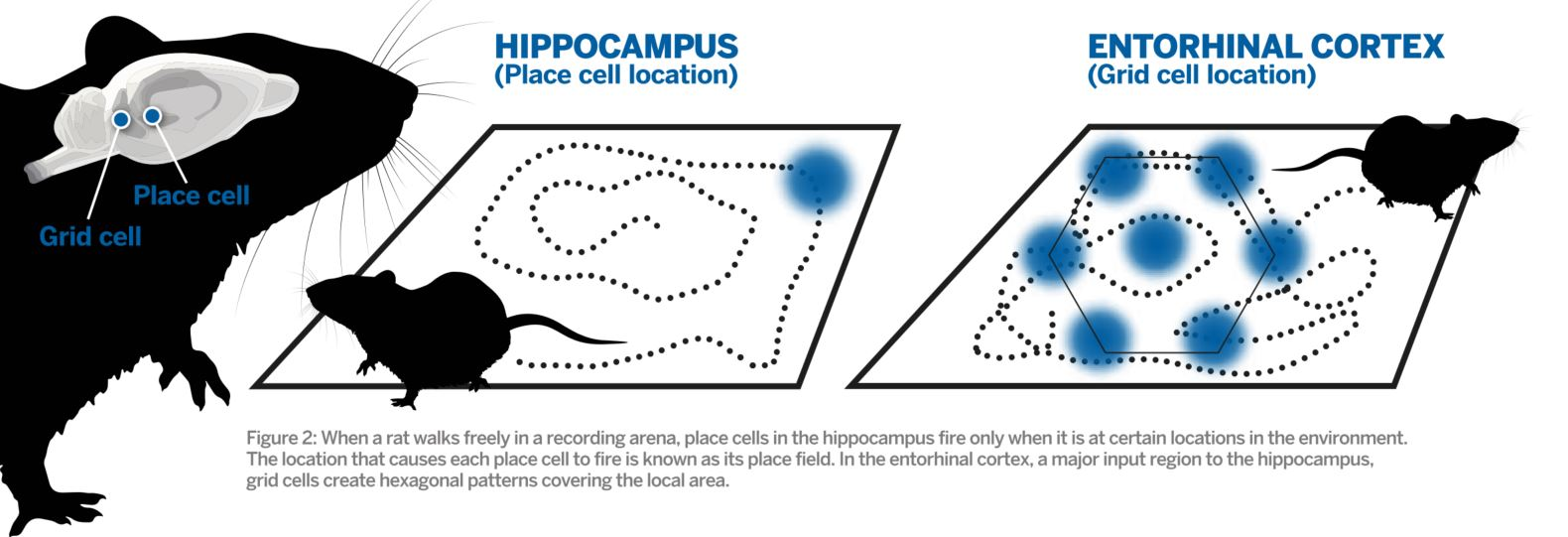
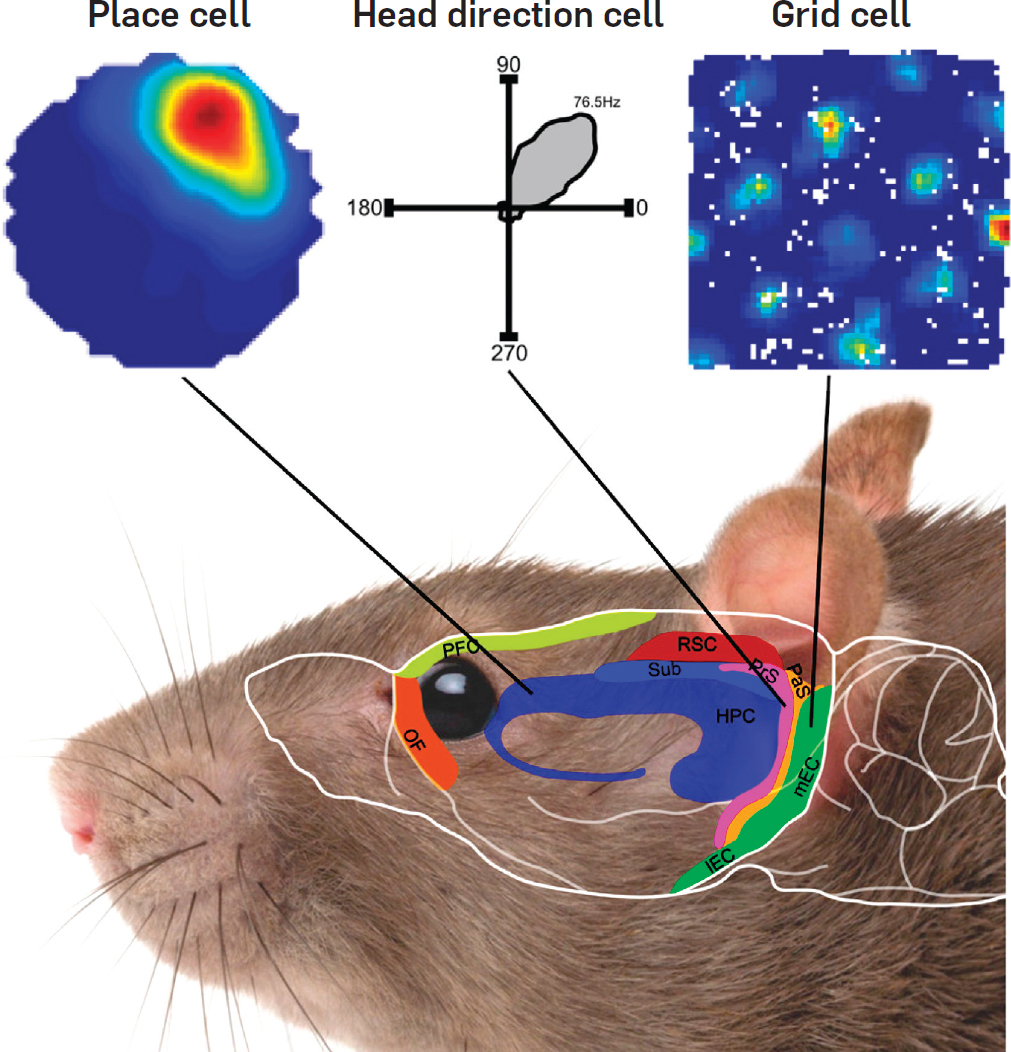
What is a place cell?
Source: Ryan Jones
- Pyramidal neuron typically located in CA1 or CA3
- Attenuate firing with changes in spatial location (one location)
- No topographic layout
- Remapping in novel environments
- Influenced by visual, tactile, olfactory, etc. cues
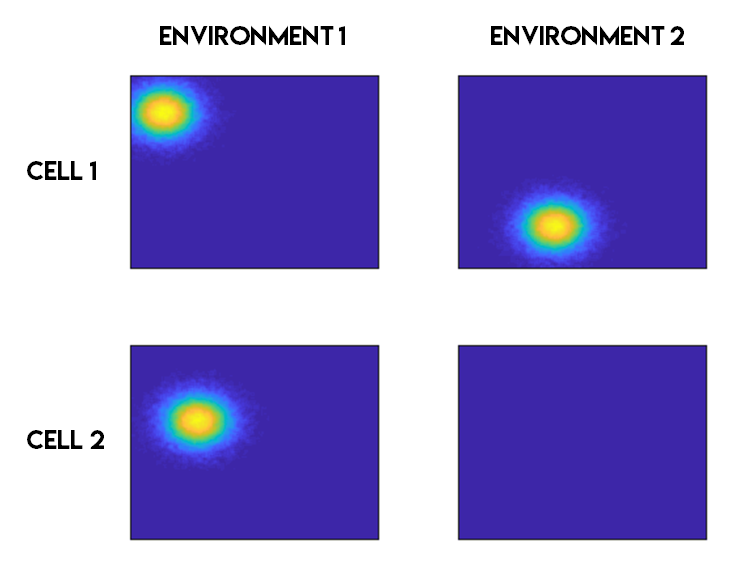
What's the difference?
Encoding in 3D
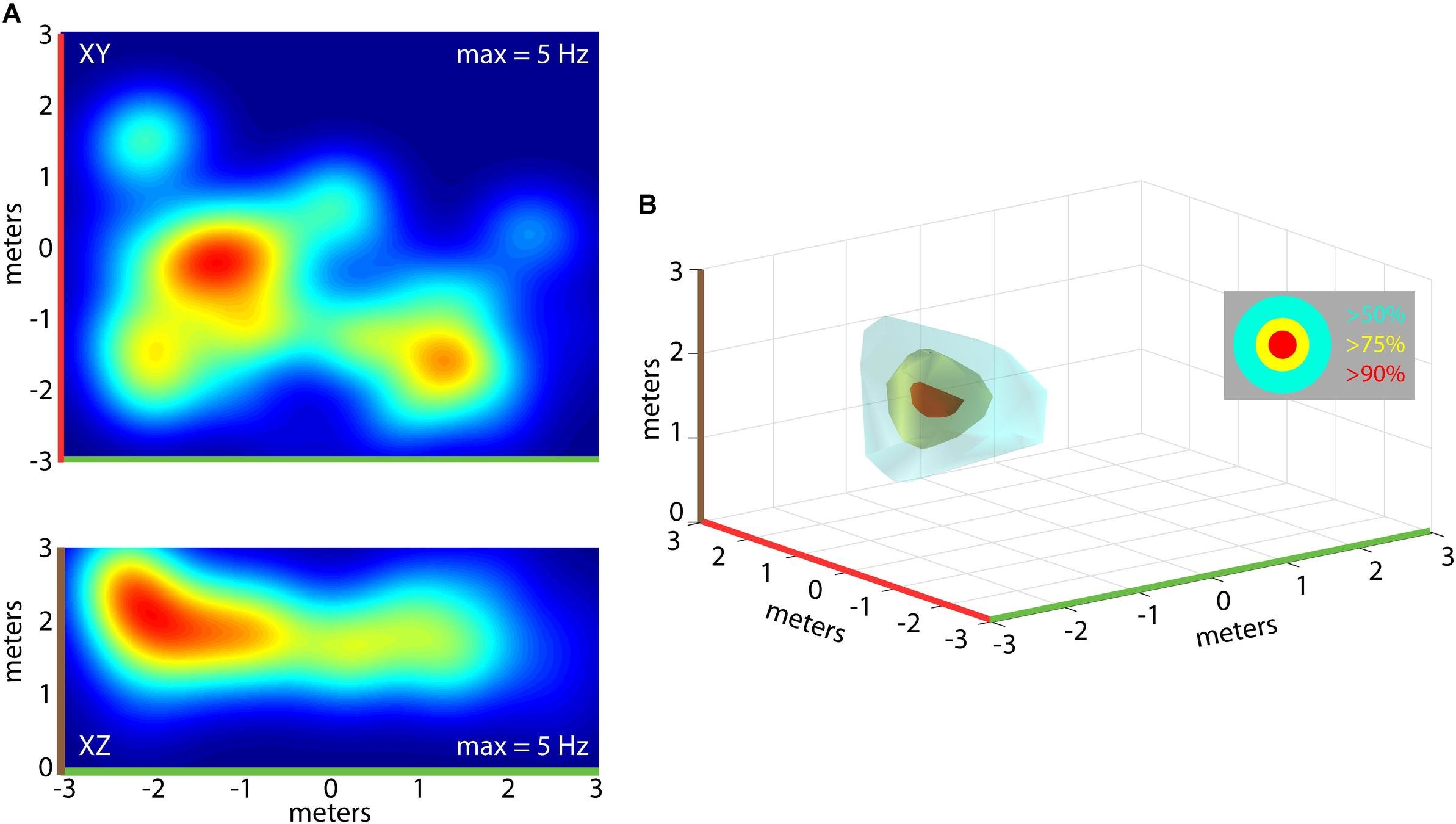
Wohlgemuth, Yu & Moss
Rhythms of the hippocampal network
Theta
Sharp-wave ripples (SWRs)
Gamma
- ~4-12 Hz
- ~150-250 Hz
- Slow ~22-55 Hz
- Fast ~60-100 Hz
Theta Oscillations
- Formation of sequential memories
- Allows brain to accept new information
- Observed during activity and REM
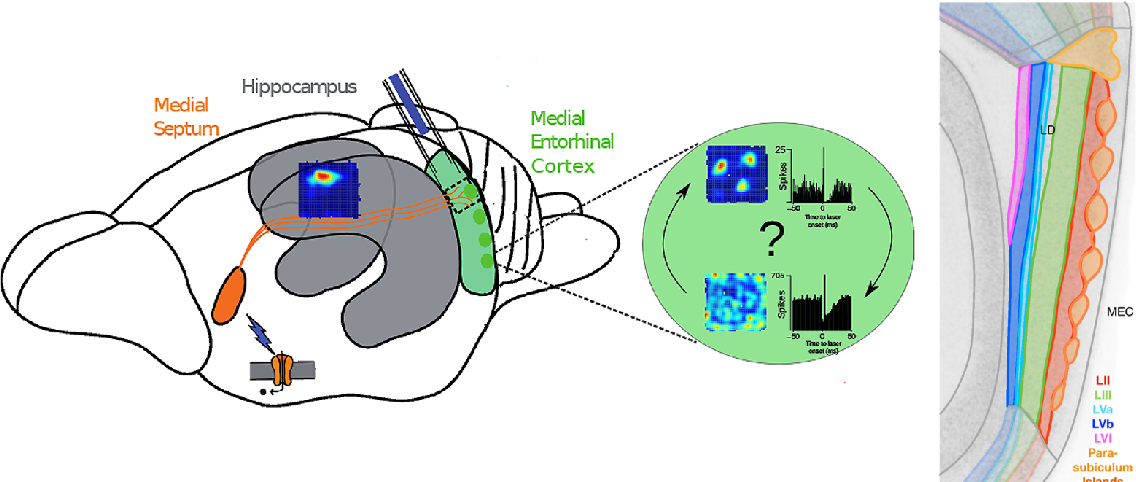
- Septum impacts firing in DG, CA1 & CA3
- Required for memory, not place fields
Theta Oscillations
- Theta sequences
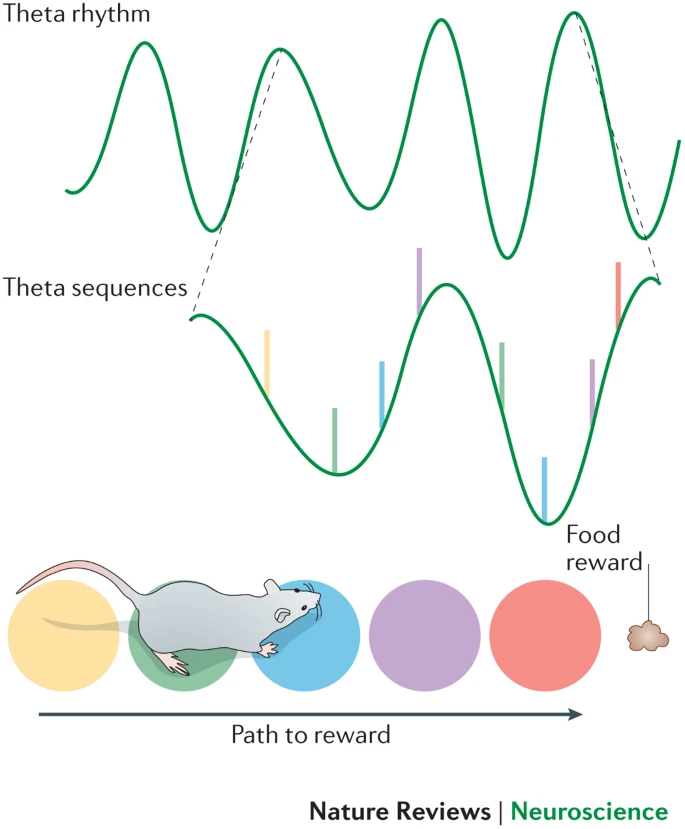
- Ordered spikes; locations traveled
- Experience-dependent, meaningful concepts
- Disruption prevents memory of sequence
Theta Oscillations
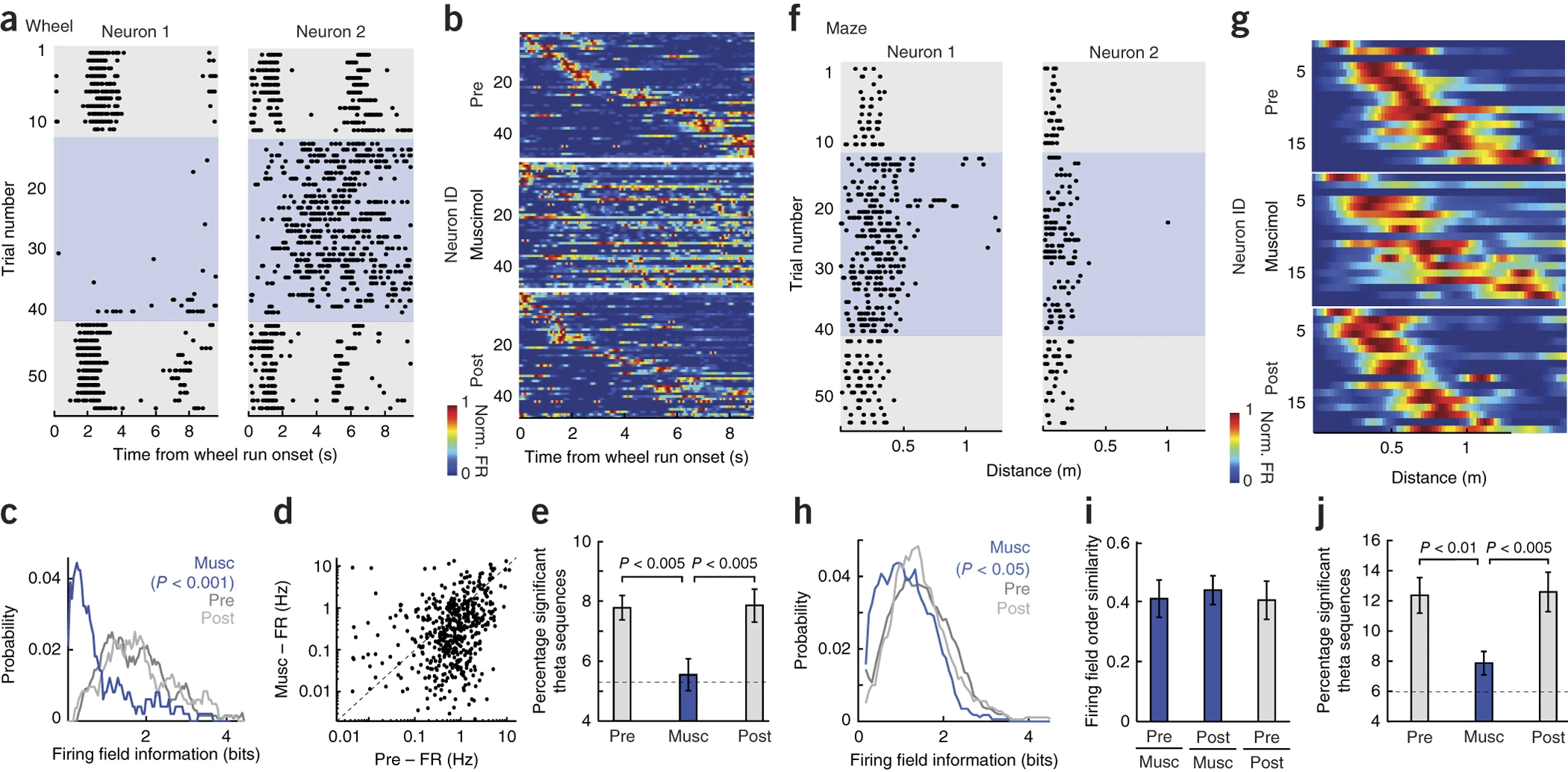
Baseline
Disruption
Return to normal
Gamma Oscillations
- Slow gamma driven by CA3, fast driven by EC
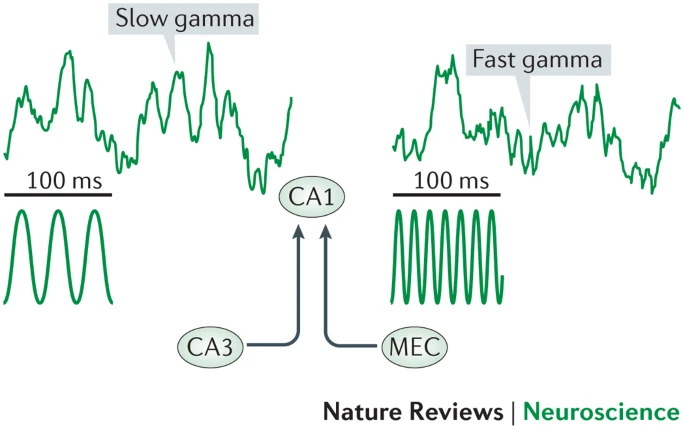
- Fast gamma related to current trajectory and recent locations
- Slow gamma retrieves spatial memories from CA3 network
- Upcoming locations when re-running
- Reflect inhibitory events in CA1, CA3 & DG
Gamma Oscillations
- Theta and gamma couple to each other
- Phase-locking observed both in awake and REM
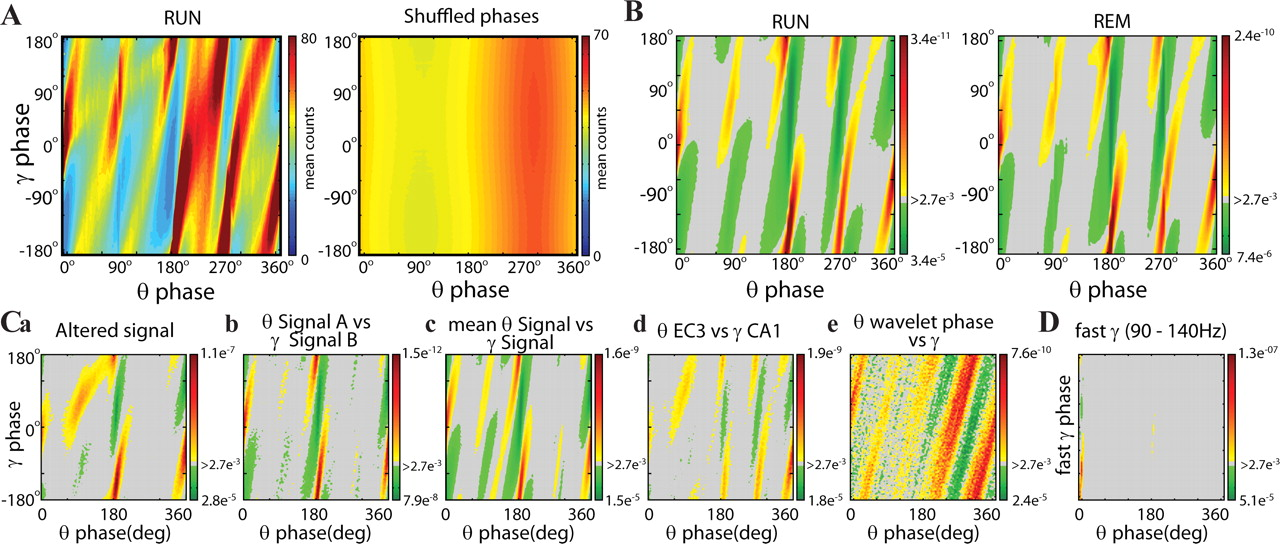
Gamma Oscillations


Sharp Wave Ripples (SWRs)
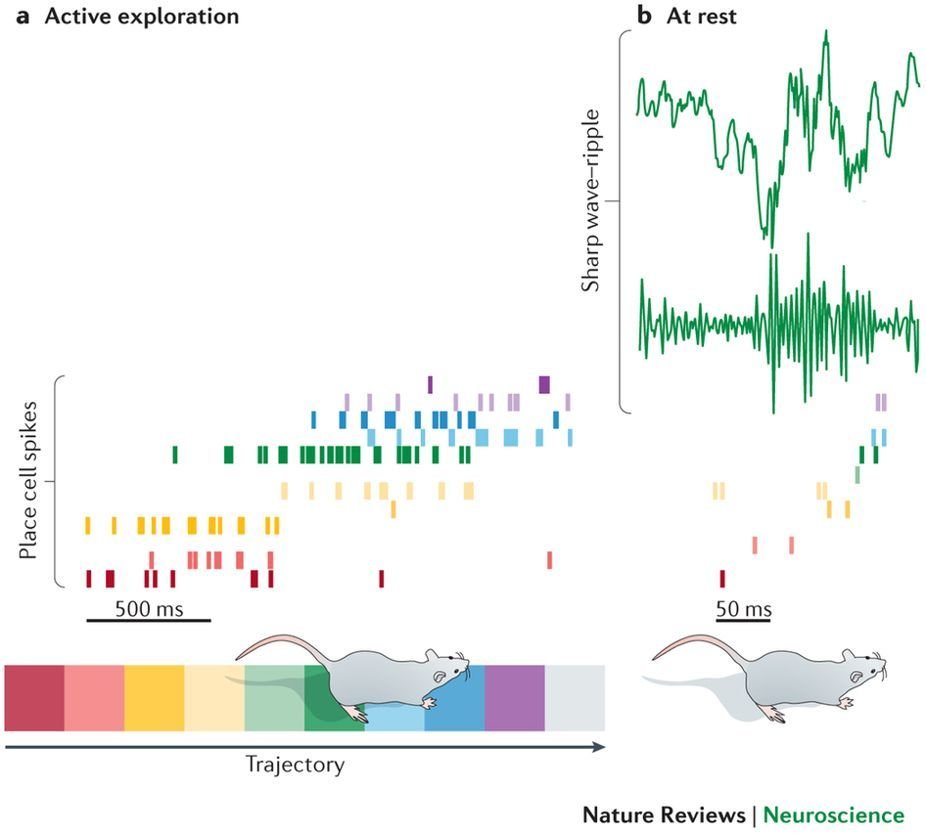
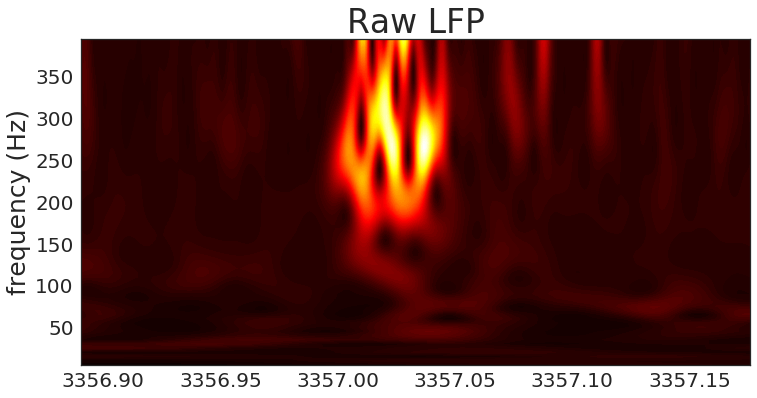
raw LFP
ripple band filtered
SWR
CA3 propagates to CA1, to produce SWRs

Adapted from Laura Colgin
Responsible for consolidation of spatial memories
def sosfiltfilt(timeseries, *, fl=None, fh=None, fs=None, inplace=False, bandstop=False,
gpass=None, gstop=None, ftype=None, buffer_len=None, overlap_len=None,
parallel=True,
**kwargs):
# make sure that fs is specified, unless AnalogSignalArray is passed in
if isinstance(timeseries, (np.ndarray, list)):
if fs is None:
raise ValueError("Sampling frequency, fs, must be specified!")
elif isinstance(timeseries, core.RegularlySampledAnalogSignalArray):
if fs is None:
fs = timeseries.fs
else:
raise TypeError('Unsupported input type!')
try:
assert fh < fs, "fh must be less than sampling rate!"
except TypeError:
pass
try:
assert fl < fh, "fl must be less than fh!"
except TypeError:
pass
if inplace:
out = timeseries
else:
out = deepcopy(timeseries)
if overlap_len is None:
overlap_len = int(fs*2)
if buffer_len is None:
buffer_len = 4194304
if gpass is None:
gpass = 0.1 # max loss in passband, dB
if gstop is None:
gstop = 30 # min attenuation in stopband (dB)
if ftype is None:
ftype = 'cheby2'
try:
if np.isinf(fh):
fh = None
except TypeError:
pass
if fl == 0:
fl = None
# Handle cutoff frequencies
fso2 = fs/2.0
if (fl is None) and (fh is None):
raise ValueError('Nonsensical all-pass filter requested...')
elif fl is None: # lowpass
wp = fh/fso2
ws = 1.4*fh/fso2
elif fh is None: # highpass
wp = fl/fso2
ws = 0.8*fl/fso2
else: # bandpass
wp = [fl/fso2, fh/fso2]
ws = [0.8*fl/fso2,1.4*fh/fso2]
if bandstop: # notch / bandstop filter
wp, ws = ws, wp
sos = sig.iirdesign(wp, ws, gpass=gpass, gstop=gstop, ftype=ftype, output='sos', **kwargs)
# Prepare input and output data structures
# Output array lives in shared memory and will reduce overhead from pickling/de-pickling
# data if we're doing parallelized filtering
if isinstance(timeseries, (np.ndarray, list)):
temp_array = np.array(timeseries)
dims = temp_array.shape
if len(temp_array.shape) > 2:
raise NotImplementedError('Filtering for >2D ndarray or list is not implemented')
shared_array_base = Array(ctypes.c_double, temp_array.size, lock=False)
shared_array_out = np.ctypeslib.as_array(shared_array_base)
# Force input and output arrays to be 2D (N x T) where N is number of signals
# and T is number of time points
if len(temp_array.squeeze().shape) == 1:
shared_array_out = np.ctypeslib.as_array(shared_array_base).reshape((1, temp_array.size))
input_asarray = temp_array.reshape((1, temp_array.size))
else:
shared_array_out = np.ctypeslib.as_array(shared_array_base).reshape(dims)
input_asarray = temp_array
elif isinstance(timeseries, core.RegularlySampledAnalogSignalArray):
dims = timeseries._data.shape
shared_array_base = Array(ctypes.c_double, timeseries._data_rowsig.size, lock=False)
shared_array_out = np.ctypeslib.as_array(shared_array_base).reshape(dims)
input_asarray = timeseries._data
# Embedded function to avoid pickling data but need global to make this function
# module-visible (required by multiprocessing). I know, a bit of a hack
global filter_chunk
def filter_chunk(it):
"""The function that performs the chunked filtering"""
try:
start, stop, buffer_len, overlap_len, buff_st_idx = it
buff_nd_idx = int(min(stop, buff_st_idx + buffer_len))
chk_st_idx = int(max(start, buff_st_idx - overlap_len))
chk_nd_idx = int(min(stop, buff_nd_idx + overlap_len))
rel_st_idx = int(buff_st_idx - chk_st_idx)
rel_nd_idx = int(buff_nd_idx - chk_st_idx)
this_y_chk = sig.sosfiltfilt(sos, input_asarray[:, chk_st_idx:chk_nd_idx], axis=1)
shared_array_out[:,buff_st_idx:buff_nd_idx] = this_y_chk[:, rel_st_idx:rel_nd_idx]
except:
raise ValueError(("Some epochs were too short to filter. Try dropping those first,"
" filtering, and then inserting them back in"))
# Do the actual parallellized filtering
if (sys.platform.startswith('linux') or sys.platform.startswith('darwin')) and parallel:
pool = Pool(processes=cpu_count())
if isinstance(timeseries, (np.ndarray, list)):
# ignore epochs (information not contained in list or array) so filter directly
start, stop = 0, input_asarray.shape[1]
pool.map(filter_chunk, zip(repeat(start), repeat(stop), repeat(buffer_len),
repeat(overlap_len), range(start, stop, buffer_len)),
chunksize=1)
elif isinstance(timeseries, core.RegularlySampledAnalogSignalArray):
fei = np.insert(np.cumsum(timeseries.lengths), 0, 0) # filter epoch indices, fei
for ii in range(len(fei)-1): # filter within epochs
start, stop = fei[ii], fei[ii+1]
pool.map(filter_chunk, zip(repeat(start), repeat(stop), repeat(buffer_len),
repeat(overlap_len), range(start, stop, buffer_len)),
chunksize=1)
pool.close()
pool.join()
# No easy parallelized filtering for other OSes
else:
if isinstance(timeseries, (np.ndarray, list)):
# ignore epochs (information not contained in list or array) so filter directly
start, stop = 0, input_asarray.shape[1]
iterator = zip(repeat(start), repeat(stop), repeat(buffer_len),
repeat(overlap_len), range(start, stop, buffer_len))
for item in iterator:
filter_chunk(item)
elif isinstance(timeseries, core.RegularlySampledAnalogSignalArray):
fei = np.insert(np.cumsum(timeseries.lengths), 0, 0) # filter epoch indices, fei
for ii in range(len(fei)-1): # filter within epochs
start, stop = fei[ii], fei[ii+1]
iterator = zip(repeat(start), repeat(stop), repeat(buffer_len),
repeat(overlap_len), range(start, stop, buffer_len))
for item in iterator:
filter_chunk(item)
if isinstance(timeseries, np.ndarray):
out[:] = np.reshape(shared_array_out, dims)
elif isinstance(timeseries, list):
out[:] = np.reshape(shared_array_out, dims).tolist()
elif isinstance(timeseries, core.RegularlySampledAnalogSignalArray):
out._data[:] = shared_array_out
return outNelpy Filtering
Why do we need prediction?
- Missing approx. 40-60% of the SWR
- Never interrogated ripple generation
- Validation of in vivo interaction
- Research context
- Interact with memory recall & consolidation
- Analyze impact on decision-making
Objective
Design Challenges
Goal: Apply machine learning techniques towards building a realtime closed loop SWR detection & disruption system.
-
Anomalous event
- Ripples happen rarely
- Complex classification problem
-
Loss value
- Weighting loss misclassifications
- Custom function
- Must be realtime applicable
Spectral Domain
Sensitivity
Convolutional vs. Recurrent
Convolutional Architecture
Multi-Layer Perceptron

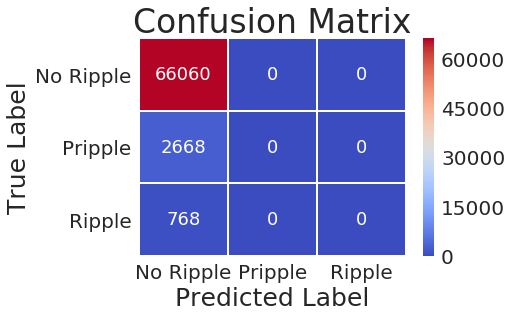

CA1
CA2
CA3
Convolutional Architecture
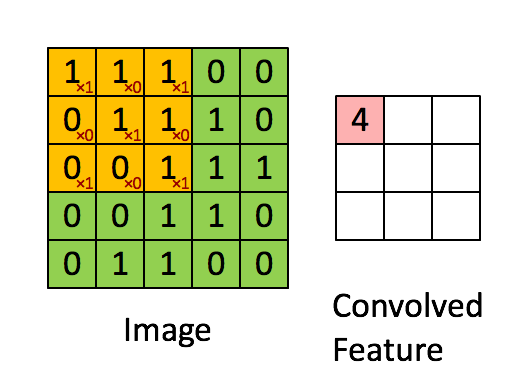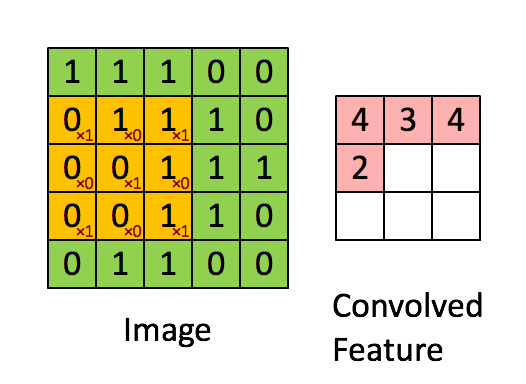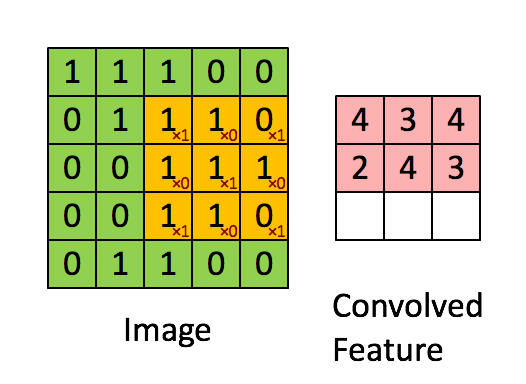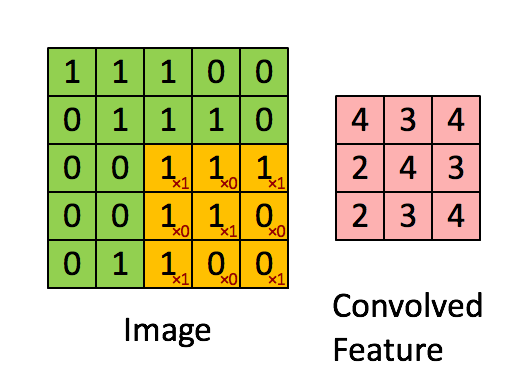
How will we approach this?
- Boosting & Cascade Learning
- Multiple mediocre classifiers
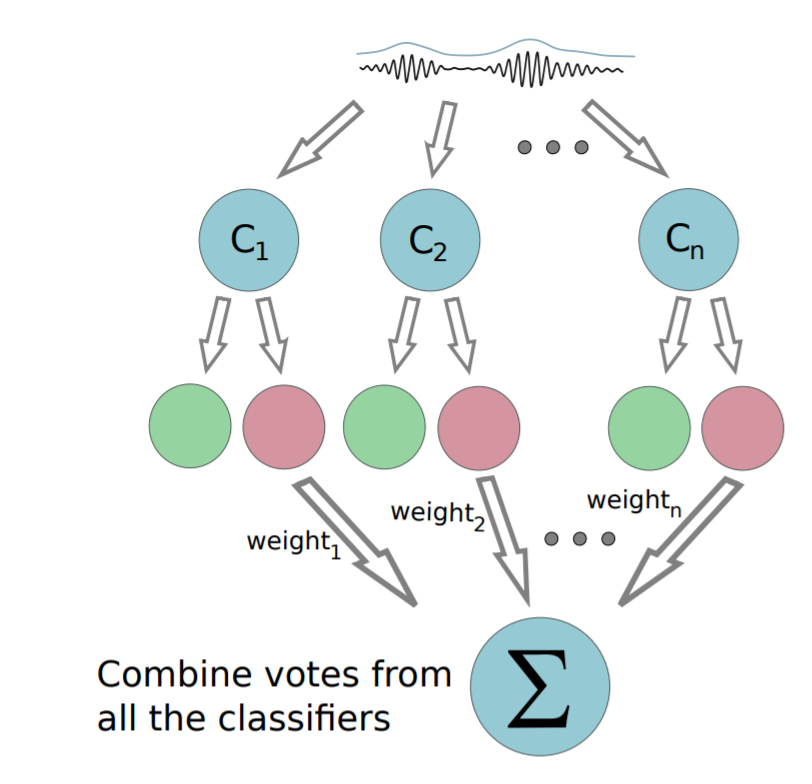
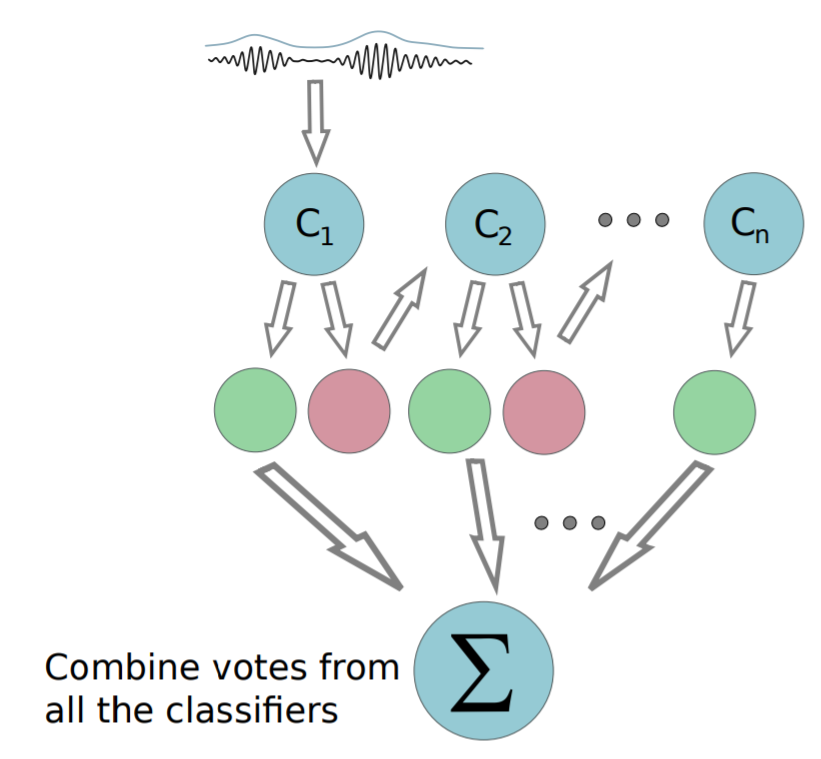
Why do we care?
-
Memory Enhancement
- Cortical p-waves & theta
-
Greater Understanding of Consolidation
- How do we improve learning?
- Neural basis of memory disorders
-
Post-Traumatic Stress Disorder
- Remove access to specific memories