
16 septembre 2016
MISSION
To offer an
automated pipeline to process diffusion neuroimaging data
- Easy to use (to convert, to run the pipeline)
- Well documented (English and French version)
- http://unf-montreal.ca/toad/html/en/index.html
- Web based QA
10 labs have used TOAD during its first year

- Conversion
- Analysis
- QA
- Visualization

Updates
- Works with Skyra scanner (soon with Prisma)
- Get a lot more information from dicom header saved in your config.cfg file

- Single command line -
dcm2toad unf-data.tar.gzConfig.cfg

[prefix]
b0_ap = b0_ap
anat = anat
dwi = dwi
grad = dwi
mag = mag
phase = phase
[correction]
echo_spacing = 0.689982557241
phase_enc_dir = 1
epi_factor = 96.0
echo_time_mag1 = 4.92
echo_time_mag2 = 7.38
[denoising]
number_array_coil = 4[methodology]
manufacturer = SIEMENS
magneticfieldstrength = 3
mrmodel = TrioTim
t1_tr = 2300.0
t1_te = 2.98
t1_ti = 900.0
t1_flipangle = 9.0
t1_voxelsize = [1.0, 1.0, 1.0]
t1_matrixsize = [256, 256]
t1_numberslices = 176
t1_fov = 256.0
t1_studyuid = 1.3.12.2.1107.5....
dwi_tr = 9300.0
dwi_te = 94.0
dwi_flipangle = 90.0
dwi_voxelsize = [2.0, 2.0, 2.0]
dwi_matrixsize = [96, 96]
dwi_numberslices = 64
dwi_fov = 192.0
dwi_studyuid = 1.3.12.2.1107.5.2....
dwi_bvalue = 1000
dwi_numdirections = 64
dwi_patfactor = 1.0
intrasession = False
[prefix]
b0_ap = b0_ap
anat = anat
dwi = dwi
grad = dwi
mag = mag
phase = phase
[correction]
echo_spacing = 0.689982557241
phase_enc_dir = 1
epi_factor = 96.0
echo_time_mag1 = 4.92
echo_time_mag2 = 7.38
[denoising]
number_array_coil = 4
- Single command line -

toad .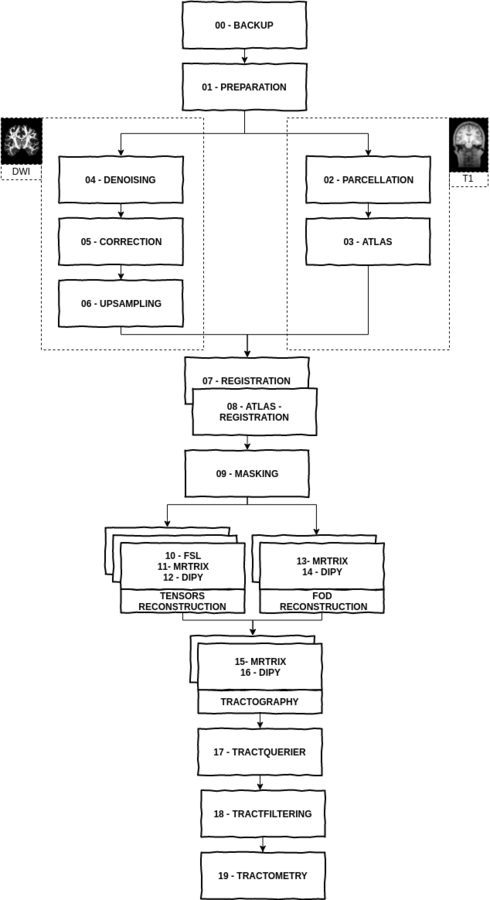

In development


In development
More options


*
In development
More options
New task

New Tasks
- TractQuerier (Wassermann et al. 2016)
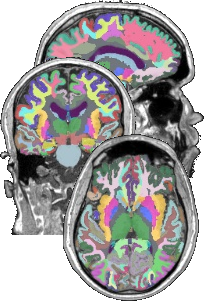
Dictionary
Atlas
Caudate.left |= 11
Caudate.right |= 50
Putamen.right |= 51
Pallidum.right |= 52
Hippocampus.right |= 53
Amygdala.right |= 54
Insula.right |= 55Dictionary
Caudate.left |= 11
Caudate.right |= 50
Putamen.right |= 51
Pallidum.right |= 52
Hippocampus.right |= 53
Amygdala.right |= 54
Insula.right |= 55Queries
cc_superiotemporal =
superiortemporal.left
and superiortemporal.right
not in subcortical.left
not in subcortical.right
not in cerebellum
- TractQuerier (Wassermann et al. 2016)


New Tasks
- TractFiltering (Cousineau et al 2016. ISMRM, Coté et al 2015. ISMRM)
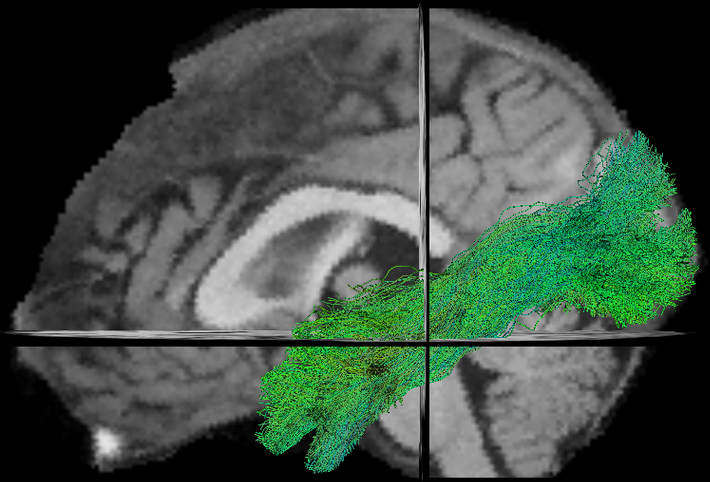
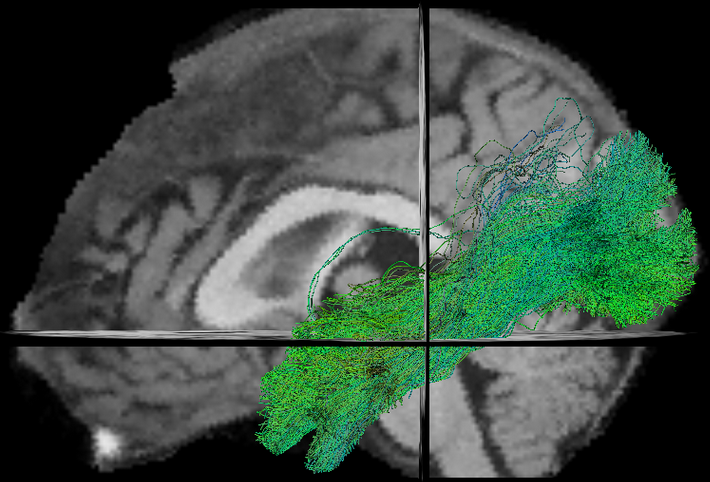
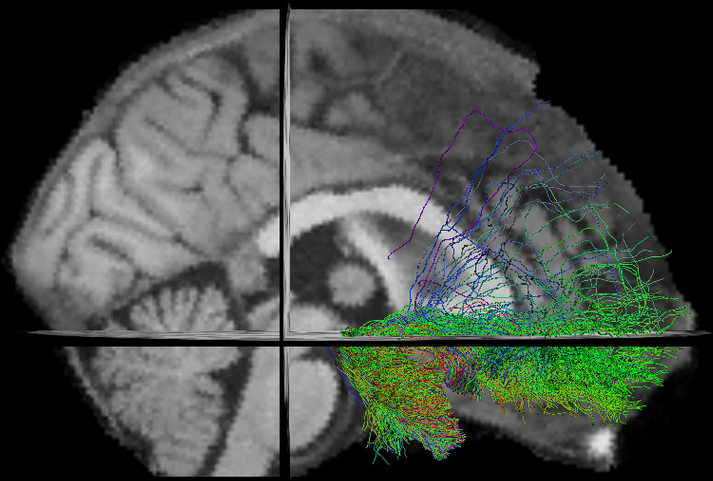
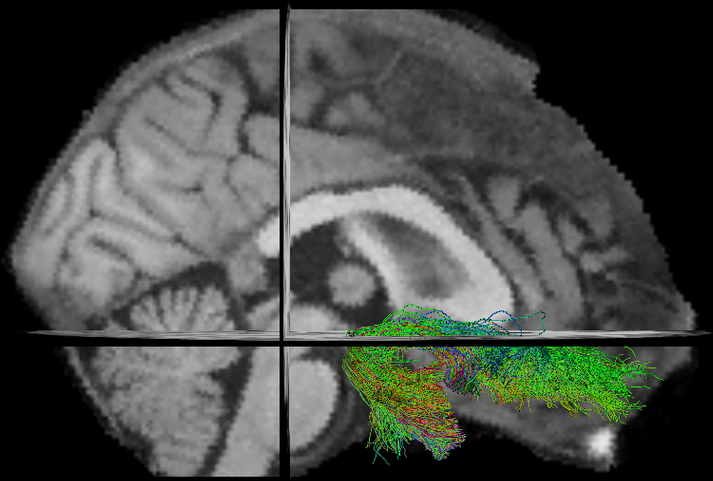

New Tasks
- Tractometry (Cousineau et al 2016. ISMRM)

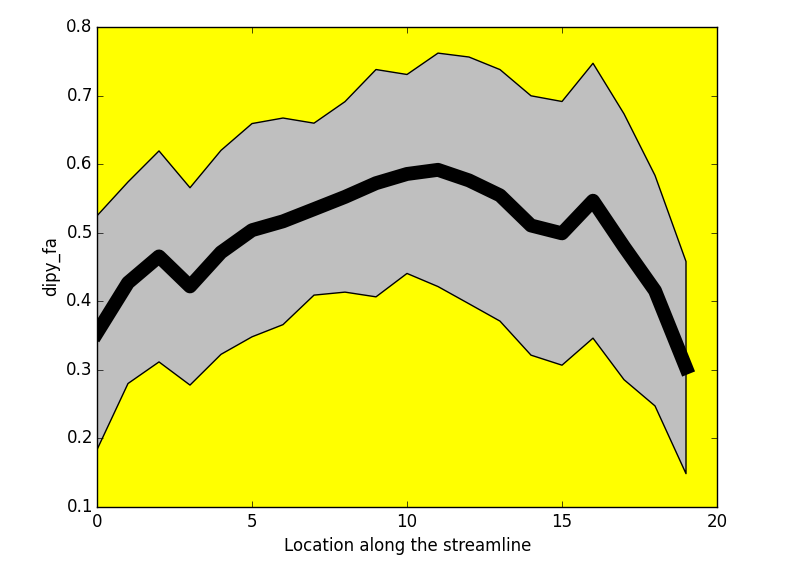
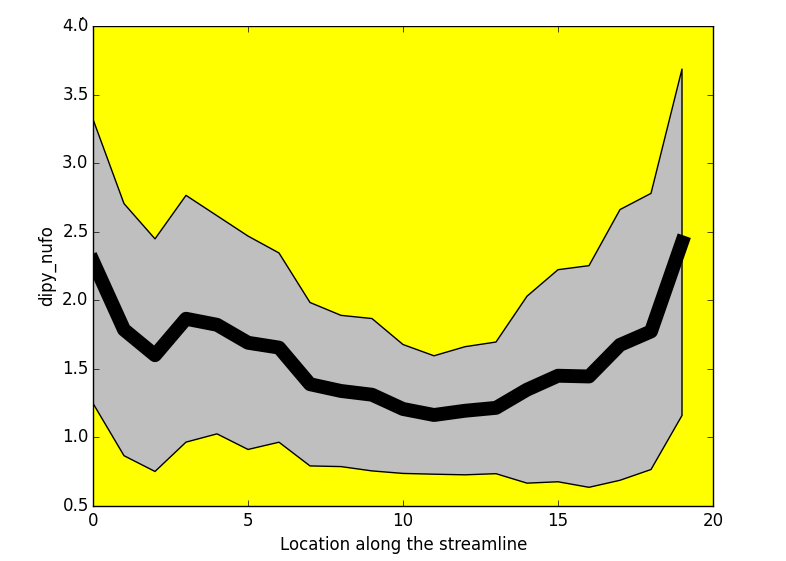
FA
NUFO
*
Metrics
- Mean value per bundle
- Mean value per point
- Profile
- Volume




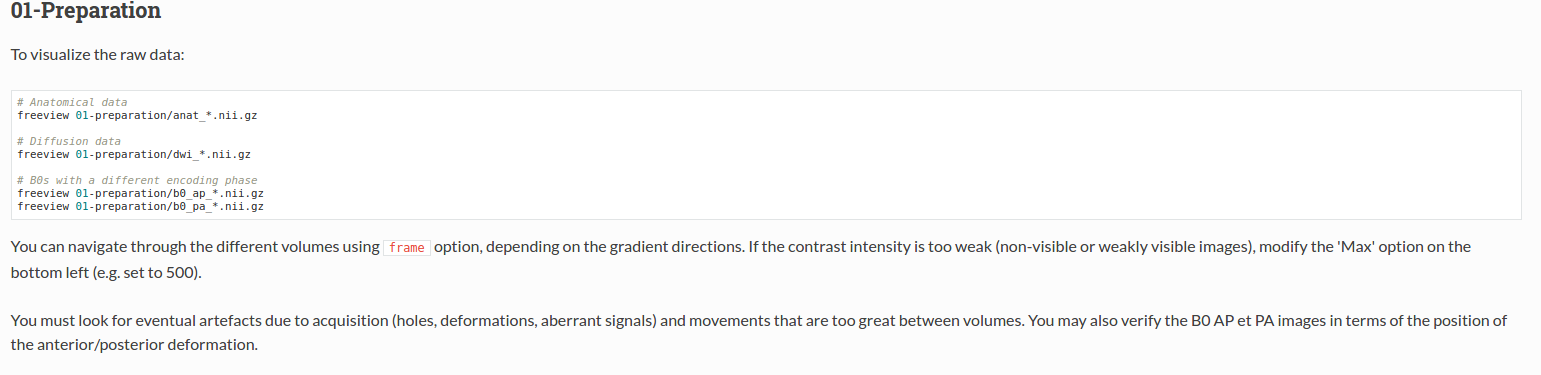
For each step of the pipeline:

----------------
REAL LIFE

-----------------
REAL LIFE
Real study with real subjects:
1 - Run dcm2toad
2 - Run toad
4 - Run Analysis
3 - Check QA

-----------------
REAL LIFE
Real study with real subjects:
4 - Run Analysis
TBSS analysis (whole brain, skeleton)
Tractography analysis (bundle specific)
Be careful with interpretations
Need to learn how to write your own queries, dictionnary
Choose a different atlas or modify wmparc
There is more than 15-TractographyMrtrix output

-----------------
REAL LIFE
Real study with real subjects:
4 - Run Tractography analysis
Need to learn how to run tractography

-----------------
REAL LIFE
-current projects-
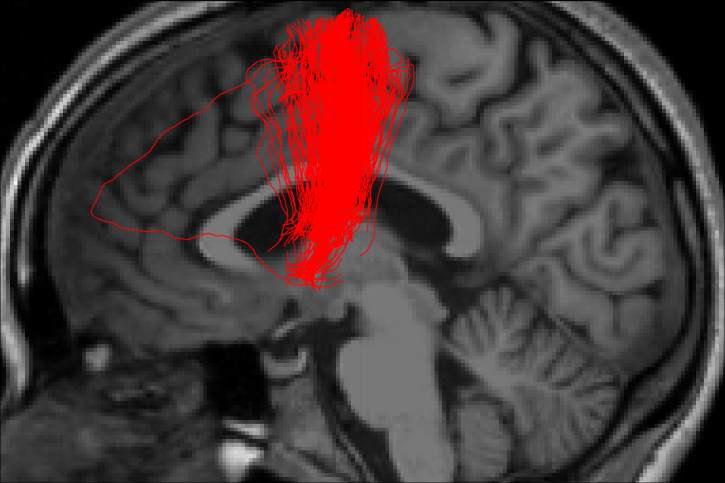
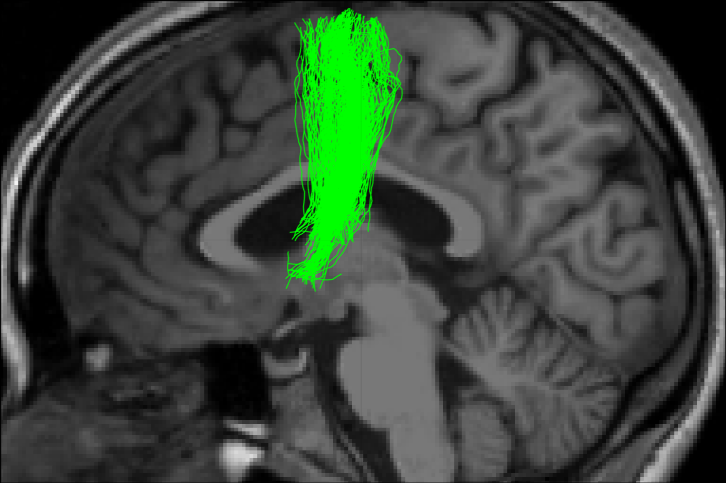

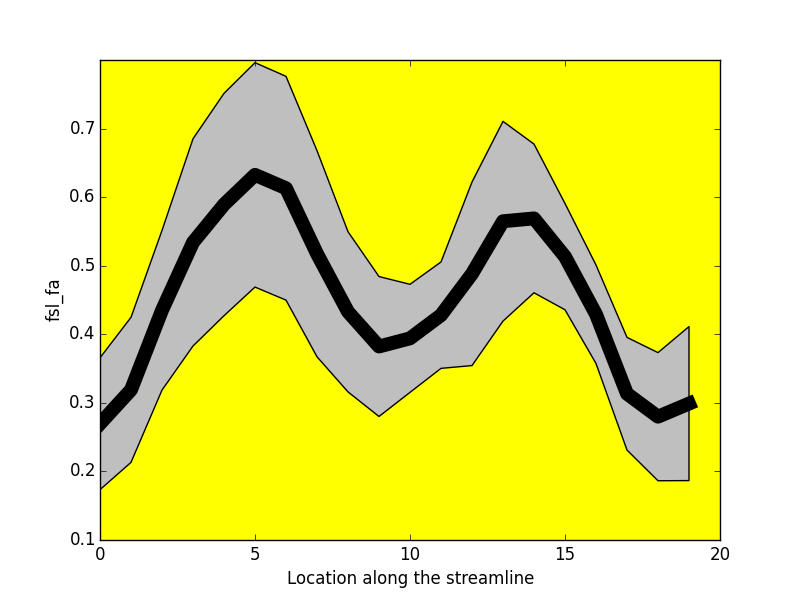
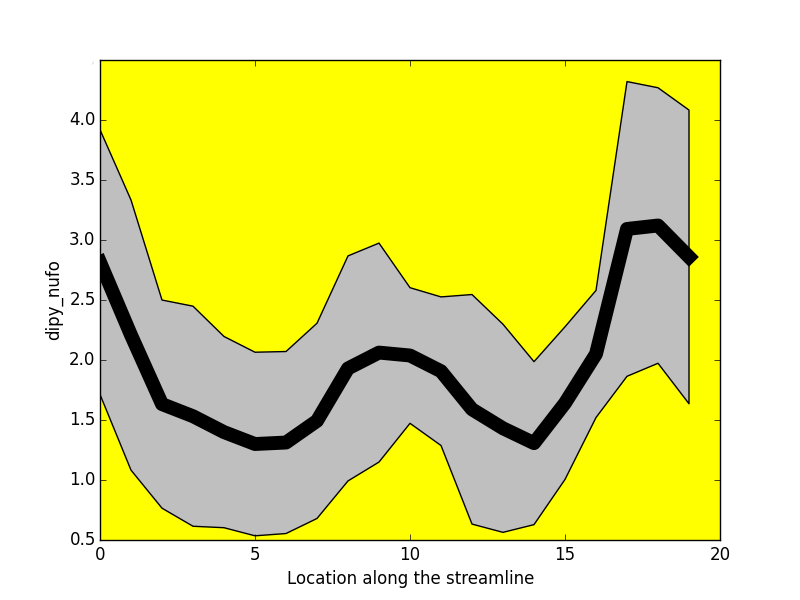
-----------------
REAL LIFE
Project: Catherine Vien, Julien Doyon
FA
NUFO
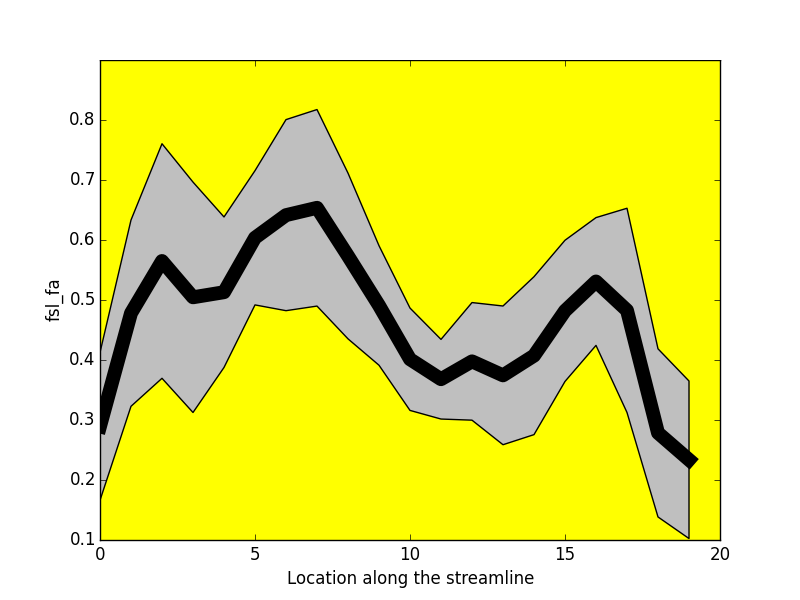
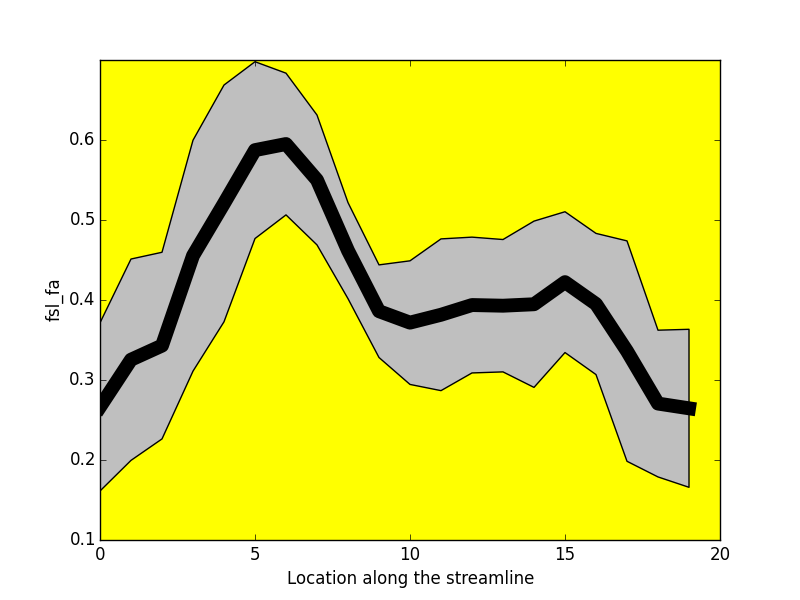
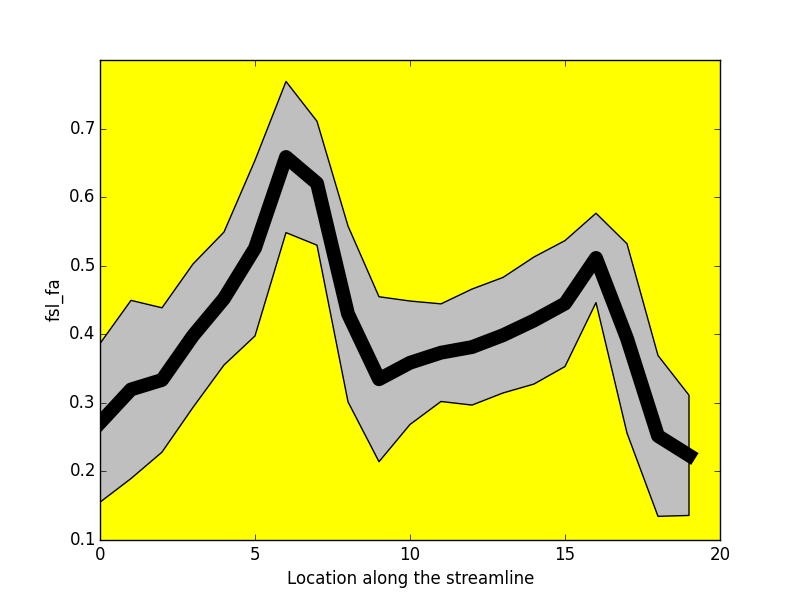
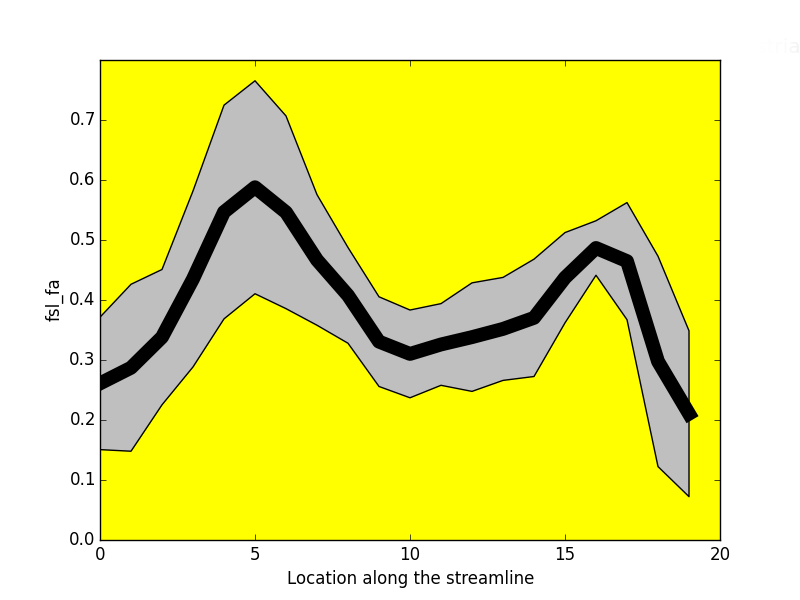


-----------------
REAL LIFE
Project: Catherine Vien, Julien Doyon

-----------------
REAL LIFE
Project: Mariem Boukadi, Simona Brambati et Maxime Descoteaux
Le rôle de la régénération des fibres de matière blanche dans la récupération spontanée du langage dans l’aphasie post-AVC

----------------
WHAT'S NEXT ?

-----------------
WHAT'S NEXT ?
- New denoising methods
- New reconstructions using multi-shells acquisitions
- New tractography algorithms (Global tractography)
- BIDS compatibility (Brain Imaging Data Structure)
- ...

MERCI
http://unf-montreal.ca/toad/html/en/index.html


toadunf.criugm@gmail.com