TractoFlow

TractoFlow
a pipeline to process diffusion MRI from RAW to tractography

our needs
-
Easy to use
-
Reproducible
-
Scalable
-
Easy to adapte
-
Easy to share
}
- Pipeline engine
- Container

- Almost like writing a program
- Very flexible
- Pipeline management built in
- Supports reproducibility (singularity, logging built in)
- Automatic parallelization
- Can reuse most of bash scripts
- Light
pipeline engine
process Denoise_DWI {
cpus params.processes_denoise_dwi
input:
set sid, file(dwi) from dwi_for_denoise
output:
set sid, "${sid}__dwi_denoised.nii.gz" into\
dwi_for_eddy,
dwi_for_topup,
dwi_for_eddy_topup
script:
"""
MRTRIX_NTHREADS=$task.cpus
dwidenoise $dwi dwi_denoised.nii.gz -extent $params.extent
fslmaths dwi_denoised.nii.gz -thr 0 ${sid}__dwi_denoised.nii.gz
"""
}container

standalone file containing everything needed to run a program or system
- Hard to exactly reproduce results after a few months
- Software versions
- Dependencies
- Hard to reproduce exactly same results with collaborators
- More and more Journals require reproducibility
- Versions are fixed -

combining all together
nextflow -C tractoflow/nextflow.config run tractoflow/main.nf \
-with-singularity singularity-tractoflow.img \
--bids CCNA_data-
Reproducible
-
Scalable
-
Easy to adapte
-
Easy to share
-
Easy to use
TractoFlow

Inputs
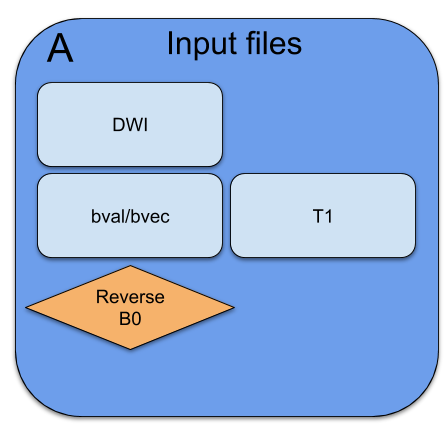
or BIDS
T1 Processing
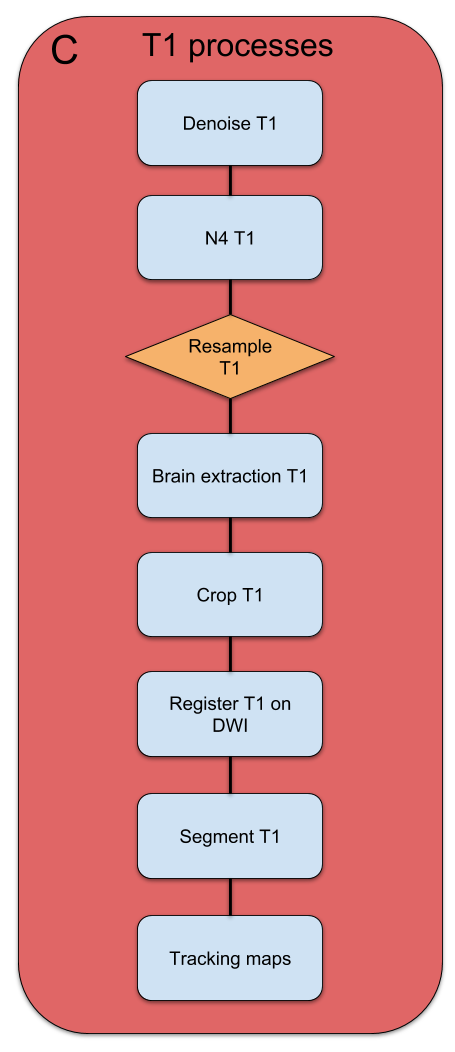
-
N4BiadFieldCorrection from ANTs
-
NLMEANS from scilpy
-
scil_resample_volume from scilpy
-
BET from FSL

T1 Processing
-
antsRegistration from ANTs
-
FAST from FSL
-
compute_maps_for_particule_filter_tracking from scilpy
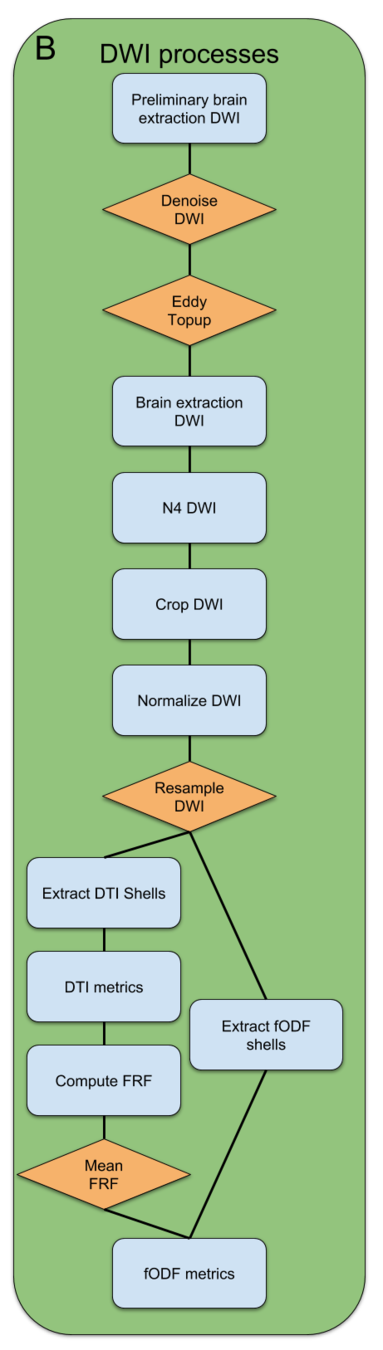
DWI Processing
-
scilpy scripts
-
dwidenoise from MRtrix
-
Eddy/Topup from FSL
-
Bet from FSL
-
N4BiadFieldCorrection from ANTs

DWI Processing
-
dwinormalise from MRtrix
-
scil_resample_volume from scilpy
-
scil_extract_dwi_shell from scilpy
-
scil_compute_dti_metrics from scilpy
-
scil_extract_dwi_shell from scilpy
-
scil_compute_mean_frf from scilpy
-
scil_compute_fodf from scilpy
-
scil_compute_fodf_metrics from scilpy
-
scil_compute_ssst_frf from scilpy
TractoFlow
B DWI processes
A Input files
C T1 processes
- DWI
- bvec/bval
- T1
Tracking
TractoFlow
Optional:
- FreeWater
- NODDI
- Freesurfer
- RecoBundle
- Tractometry + Stats
RecoBundle
Garyfallidis et al., Neuroimage, 2017.
Tractometry
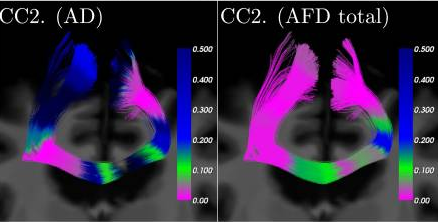
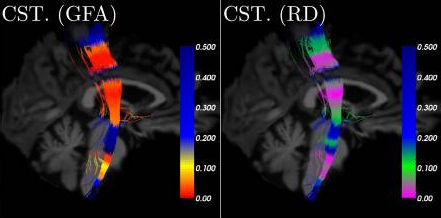
Heatmap of p-values projected on different pathways of a specific subject
Cousineau et al Neuroimage Clin. 2017
TractoFlow - Reproducibiliy
- All DWI preprocesses steps are 100% reproducible
- T1 BET and registration introduce small differences
- Maximum MAE: 0.00063
- Minimum CC: 0.99988
- Tracking reproducibility
- Seeding
the pipeline can reach 100% reproducibility with a 50% slower runtime
98% reproducibility with default parameters
3 datasets and 3 computer configurations
Thank you


M.Descoteaux
J-C.Houde


G.Theaud
F.Rheault
F. Morency


scil.usherbrooke.ca
TractoFlow

a pipeline to process diffusion MRI from RAW to tractography


based on
soon available on

using

