Reading in the Literate Ape:
Variations in the hemodynamic response associated with saccadic eye-movements during reading.

The Oddity of Man
- Reading and writing are unique to man
- It has no evolutionary basis (as far as we can tell).
- It gives us the ability to preserve and transmit knowledge across time and space.
- According to the archeological record, disparate writing systems appeared independently of each other (e.g. cuneiform, Mayan).
"And a book of remembrance was kept, in the which was recorded, in the language of Adam, for it was given unto as many as called upon God to write by the spirit of inspiration;
"And by them their children were taught to read and write, having a language which was pure and undefiled."
–Moses 6:4-6
https://www.lds.org/
Known reading network
- Ventral (orthographic) system
- VWFA
- Dorsal (phonologic) system
- Supramarginal gyrus
- Angular gyrus
- Sup. temporal cortex
- Dorsal premotor cortex
Prerequisites to reading
- Eye(s) with normal or corrected to normal vision
- Oculomotor control
- Language fluency
Types of Eye Movements
- Fixations – when the fovea is stationary on a visual target.
- Saccades – a ballistic movement in which the eye changes orientation between fixations.
Saccade Execution Network
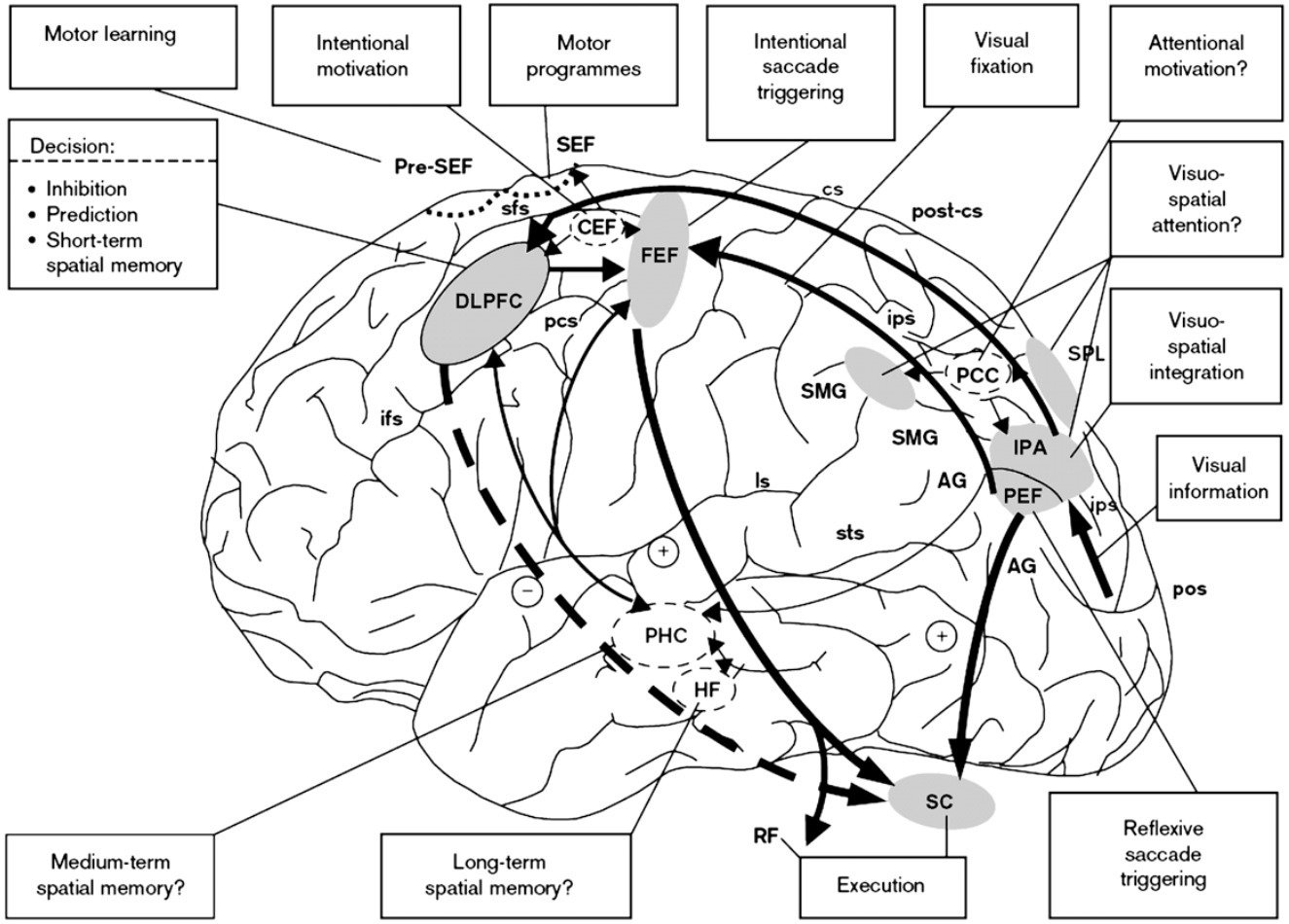
Image courtesy of: Müri, R. M., & Nyffeler, T. (2008). Neurophysiology and neuroanatomy of reflexive and volitional saccades as revealed by lesion studies with neurological patients and transcranial magnetic stimulation (TMS). Brain and Cognition, 68(3), 284-292. doi:10.1016/j.bandc.2008.08.018
The Study
- The aim of this analysis is to explore saccadic control during reading and determine ROIs necessary for saccade execution and planning for future investigation.
The Study
How we did it:
- Participants underwent 3 functional scans during which they were presented with 18 paragraphs to read per scan.
- Eye movements were recorded during functional scans.
- Structural scans were also performed.
Methods
Participants
- 43 BYU undergraduates, recruited via SONA.
- Normal or corrected to normal vision.
- No contraindications for an MRI.
Materials
- Text was taken from online news articles and literature.
Acquisition
Scanner
- Siemens 3T Trio Tim with 12-channel head coil.
Parameters
- 3 consecutive blocks of interleaved T2* weighted echo-planar imaging.
- Slice number 43
- Orientation is transverse
- Phase encoding direction is anterior to posterior
- Rotation is 0°
- FOV= 224x224
- Matrix 64x64
- Slice thickness 3.0 mm
- TR 2500 ms, TE 28 ms
- 134 repetitions
- Flip angle 90°
Acquisition
Parameters
- T1-weighted imaging
- Orientation is sagittal
- Anterior to posterior phase encoding
- FOV=218x250
- Matrix 256x256
- Slice thickness 1.00 mm
- TR 1900 ms, TE 2.26 ms
- Flip angle is 9°
Eye Tracking Methods
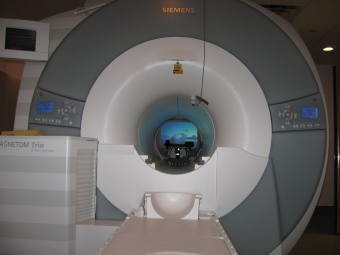
SR Research Eyelink 1000 Plus
- Spatial resolution 0.01 degrees.
- Sampling rate 1000 Hz.
- Text adjusted to 3.4 letters per degree of visual angle.
- Head movement stabilized with foam pads.
Preprocessing & Analysis
Eye tracking
- Saccades were excluded if they included a blink or if they ended outside predefined interest areas (i.e. did not land on a word).
- Fixations shorter than 50 msec or longer than 1500 msec were also excluded.
- Therefore 75,270 saccade/fixation pairs were included in the analysis.
- These were then temporally registered to the start of each functional scan.
Imaging data were preprocessed and analyzed using:
- Analysis of Functional NeuroImages
- Advanced Normalization Tools
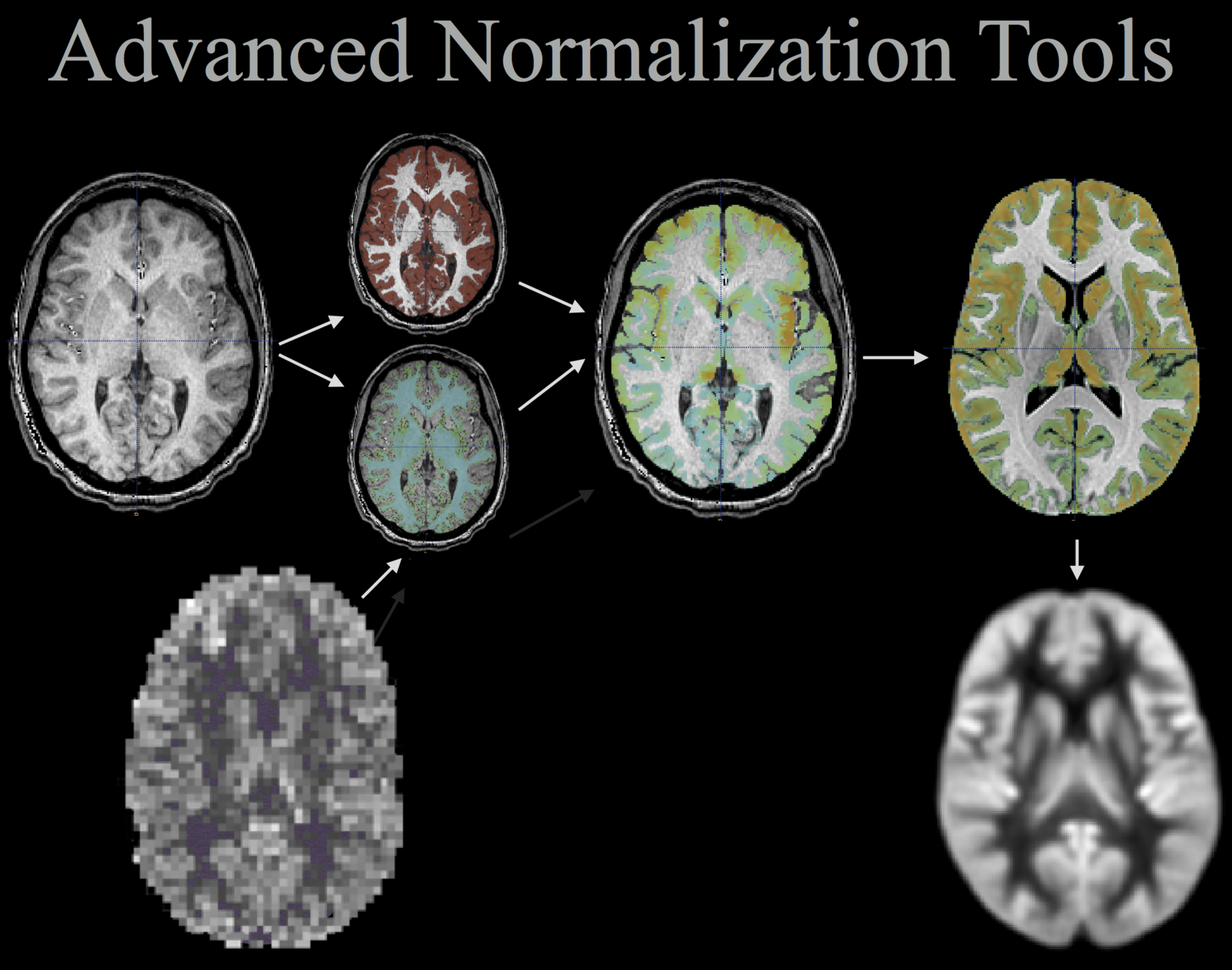

#DICOM files were imported
to3d
-time:zt <slice number> <number of TRs per run> <TR in msec> \
alt+z epi[1,2,3] <epi dicom>
to3d -prefix struct <t1 dicom>
#Co-registration of structural to functional
3dWarp -oblique_parent <epi3> \
-prefix <warped file prefix> <input t1>
#Slice time correction
3dTshift -verbose -prefix <new prefix> <input epi>
#Motion correction within runs
3dvolreg
-base <middle TR of run>
-prefix <output epi prefix>
-1Dfile <1D output file> <input slice time corrected epi>
#Motion correction across runs
3dvolreg
-base <middle TR of reference run>
-prefix <aligned epi> <input epi>
#Motion censor creation
cat <1D output file> >> motion.txt
move_censor.plPreprocessing
#Mask creation
3dSkullStrip
-input <coregistered t1> \
-o_ply <output name>
3dfractionize
-template <reference epi> \
-input <skullstripped t1> \
-prefix <output prefix>
3dcalc
-a <fractionized t1> \
-prefix <prefix>
-expr "step(a)"Mask Creation
#Regression
3dDeconvolve \
-input <epi run 1> <epi run 2> <epi run 3> \
-mask <t1 mask> \
-polort A \
-num_stimts 7 \
-stim_file 1 "motion.txt[0]" -stim_label 1 "Roll" -stim_base 1 \
-stim_file 2 "motion.txt[1]" -stim_label 2 "Pitch" -stim_base 2 \
-stim_file 3 "motion.txt[2]" -stim_label 3 "Yaw" -stim_base 3 \
-stim_file 4 "motion.txt[3]" -stim_label 4 "dS" -stim_base 4 \
-stim_file 5 "motion.txt[4]" -stim_label 5 "dL" -stim_base 5 \
-stim_file 6 "motion.txt[5]" -stim_label 6 "dP" -stim_base 6 \
-stim_times_AM2 7 <Timing file> 'dmBLOCK' \
-stim_label 7 "<label>" \
-num_glt 1 \
-gltsym 'SYM: <label>' \
-glt_label 1 <label> \
-censor 'motion_censor_vector.txt[0]' \
-nocout -tout \
-bucket <output prefix> \
-xjpeg <design JPEG> \
-jobs 2 \
-GOFORIT 12
#Output blurring
3dmerge
-prefix <output prefix> \
-1blur_fwhm 5.0 \
-doall <input file>Individual Subject Regression
#T1 transformation
ants.sh <transformation dimensions> <model template> <subject t1>
#EPI transformation
ANTifyFunctional <input structural> <model template> <blurred epi> ANTs Transformation
3dttest++ -prefix <output prefix> \
-mask <model template mask> \
-setA \
<Subject 1 transformed regression input> \
<Subject 2 transformed regression input> \
...
<Subject 43 transformed regression input> \Group t-test
Results
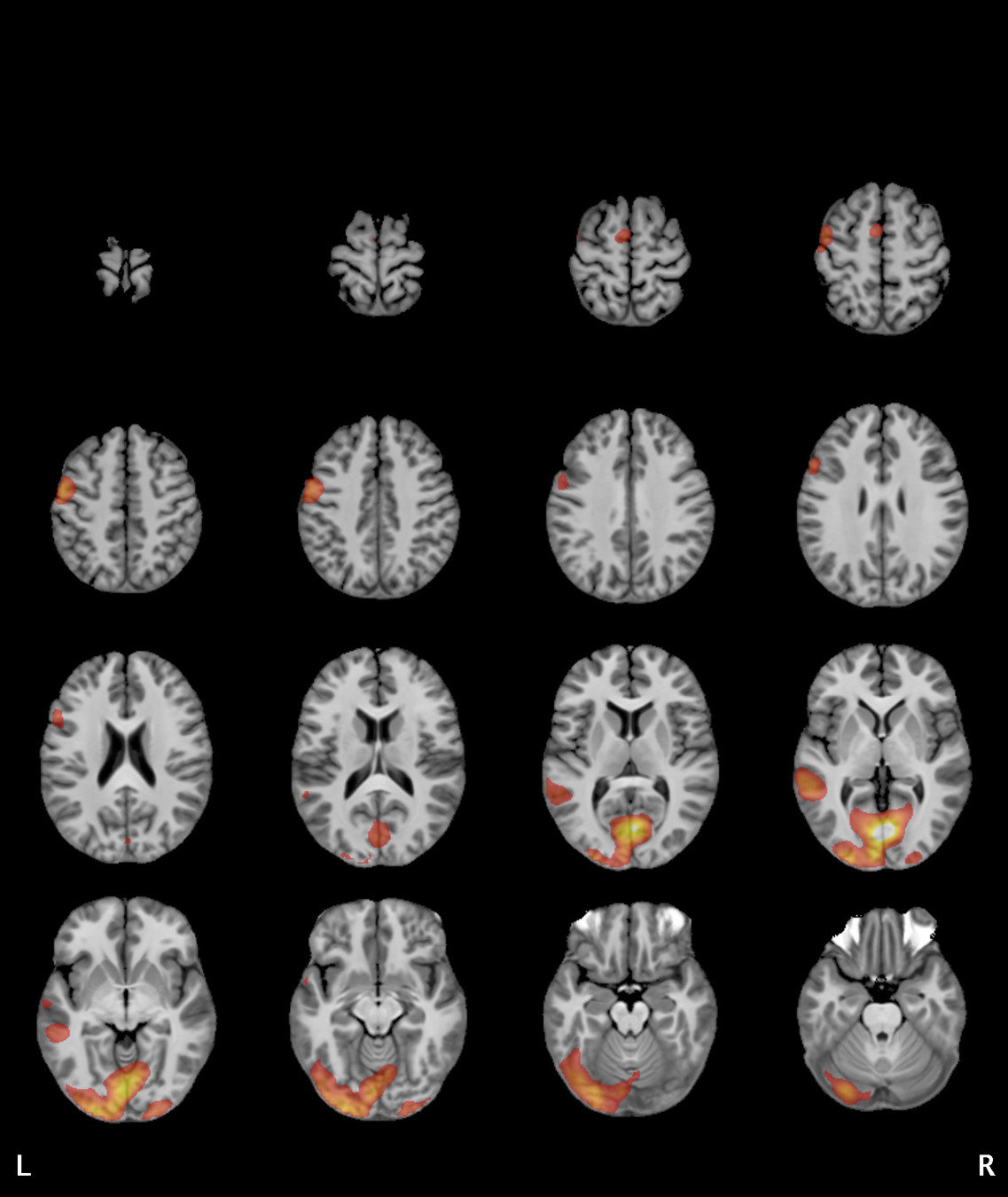
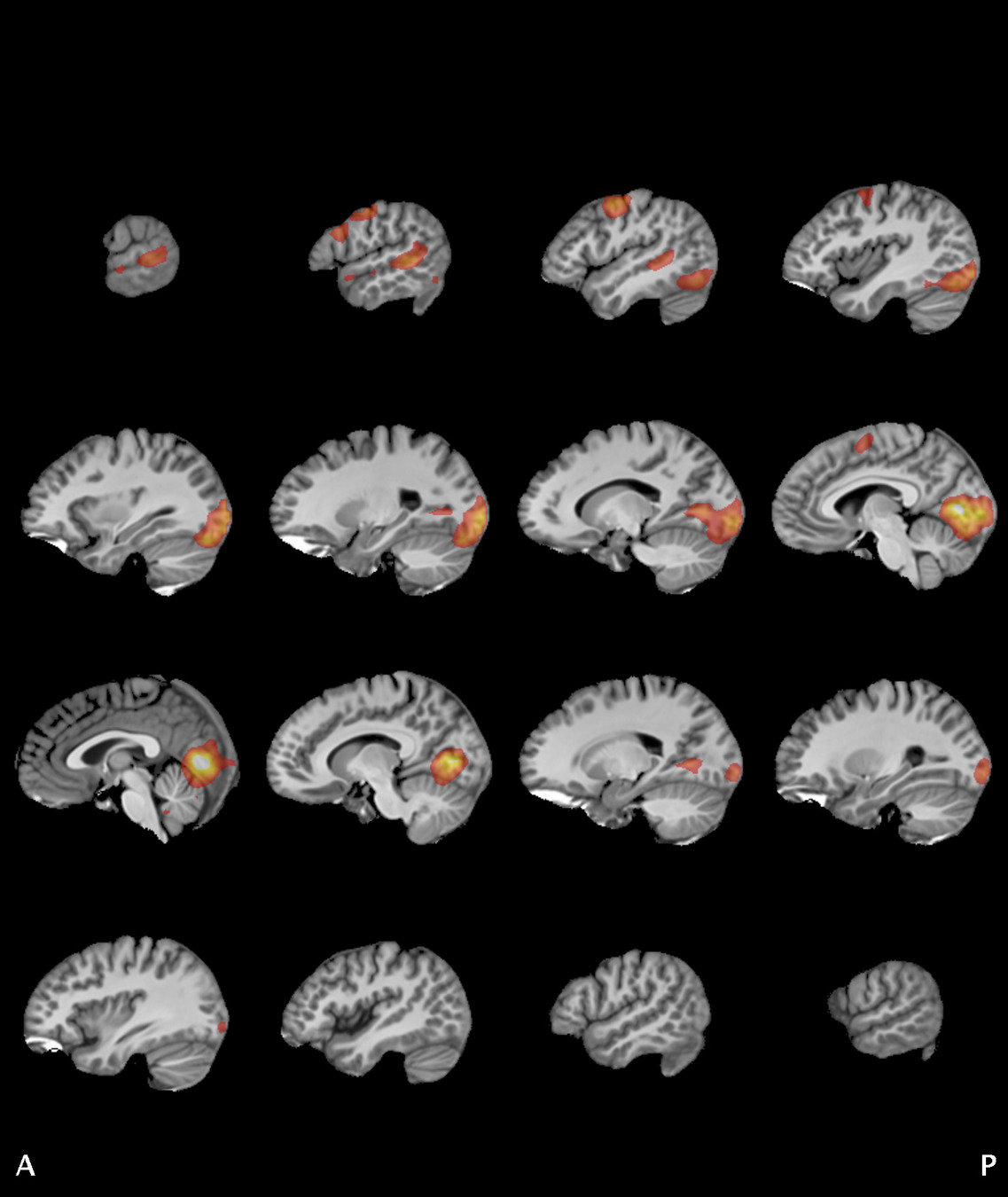
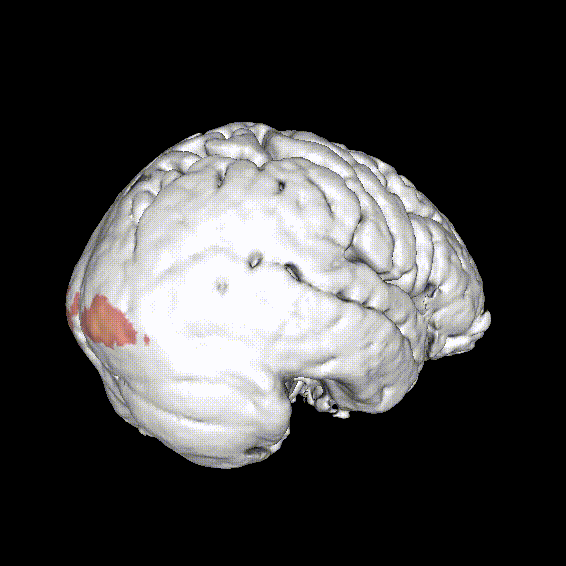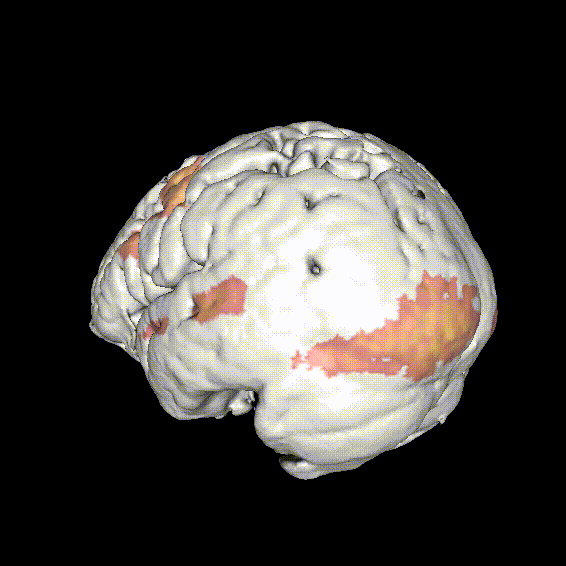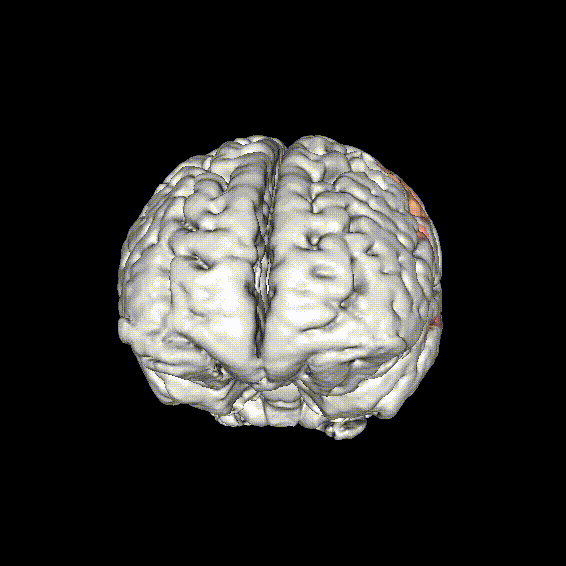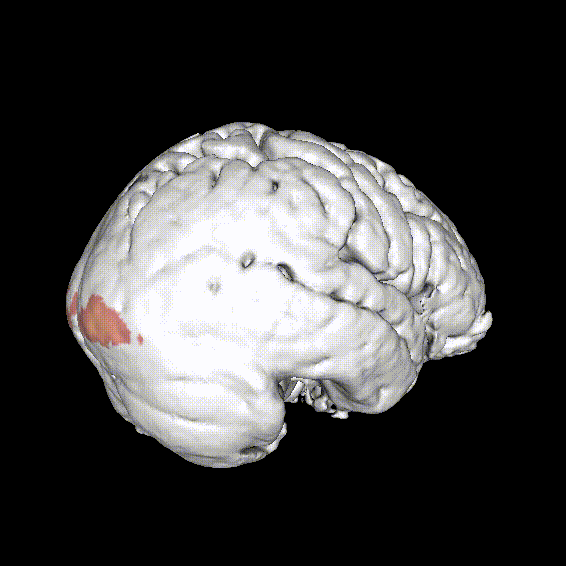
Significant ROIs
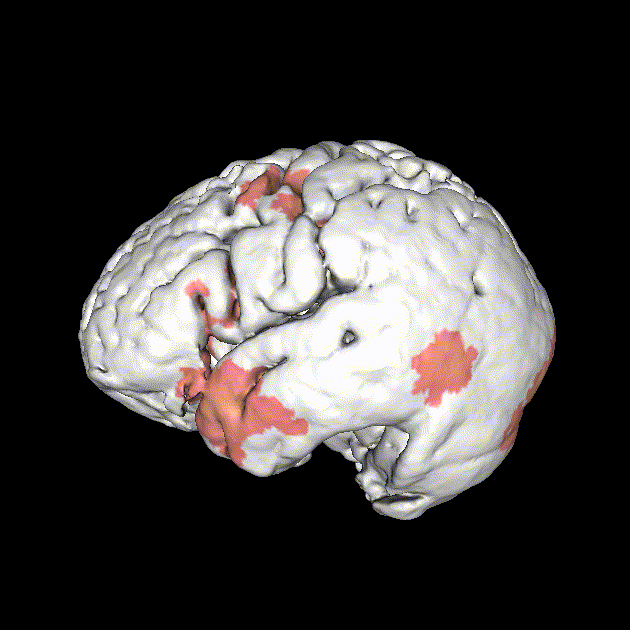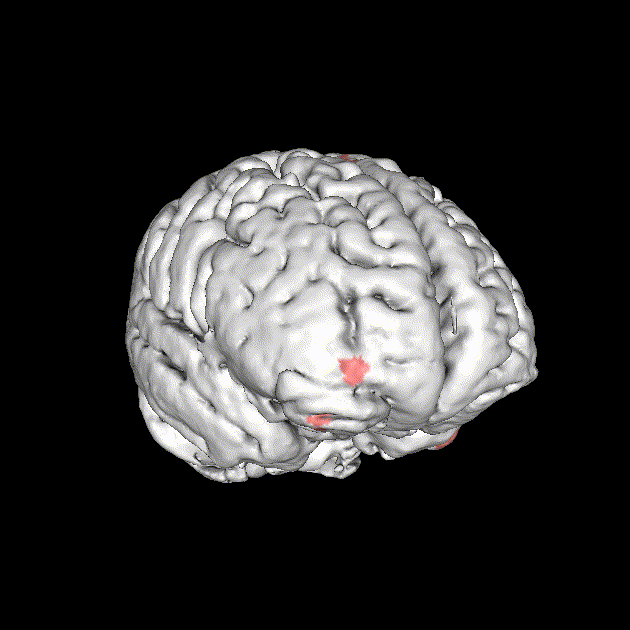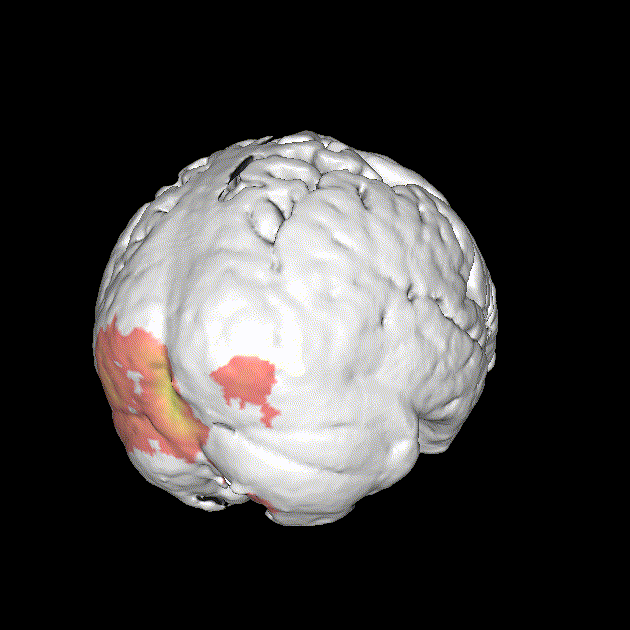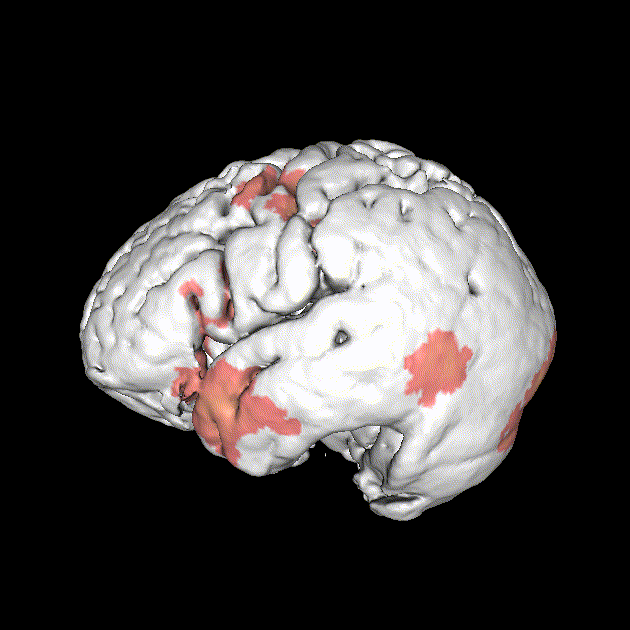
| Voxels | Peak - x | Peak - y | Peak - z | Anatomical location |
|---|---|---|---|---|
| 1923 | -1.5 | 79.5 | -9 | R/L Lingual Gyrus |
| 495 | 52.5 | -10.5 | -18.0 | L. Mid. Temporal Gyrus |
| 395 | 46.5 | 16.5 | 33.0 | L. Precentral Gyrus |
| 135 | 1.5 | 38.5 | -12.0 | L. Ant. Cerebellum |
| 56 | 7.5 | 19.5 | -3.0 | L. Thalamus |
| 55 | -22.5 | -94.5 | -9.0 | R. Inf. Occipital Gyrus |
| 55 | 52.5 | -7.5 | 15.0 | L. Inf. Frontal Gyrus |
| 54 | 4.5 | 7.5 | 60.0 | L. Medial Frontal Gyrus |
| 29 | -46.5 | -7.5 | -24.0 | R. Temporal Pole |
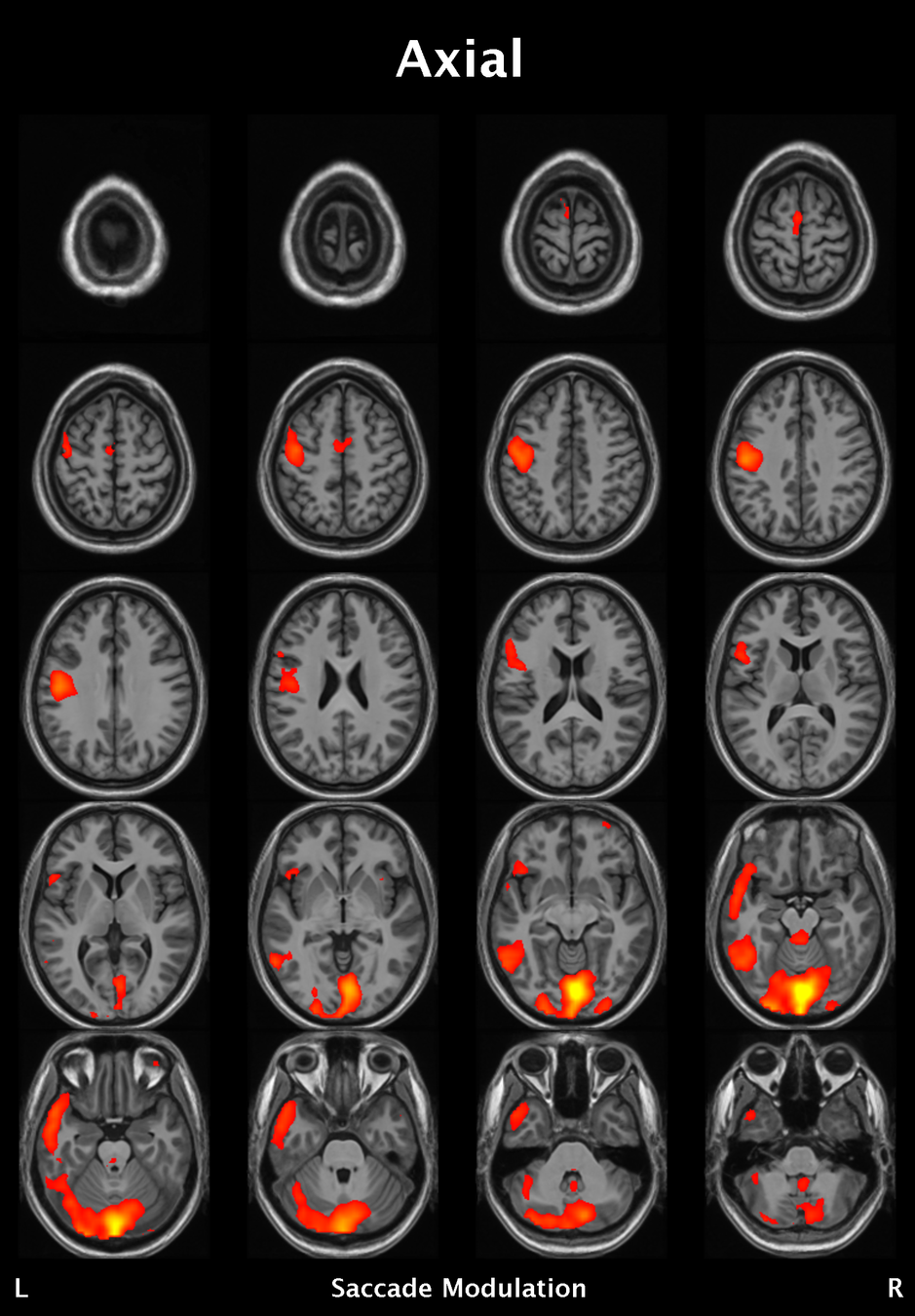
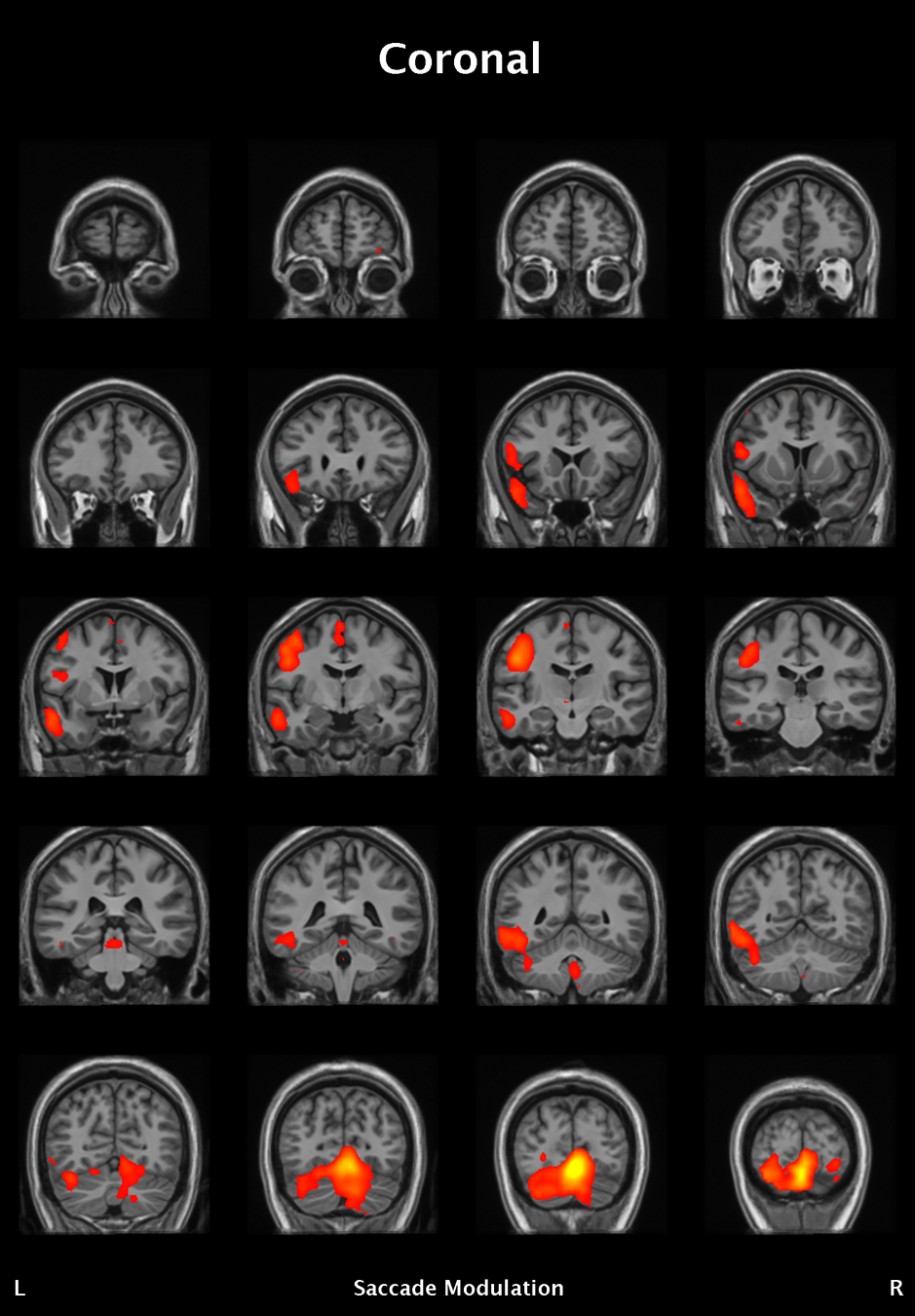
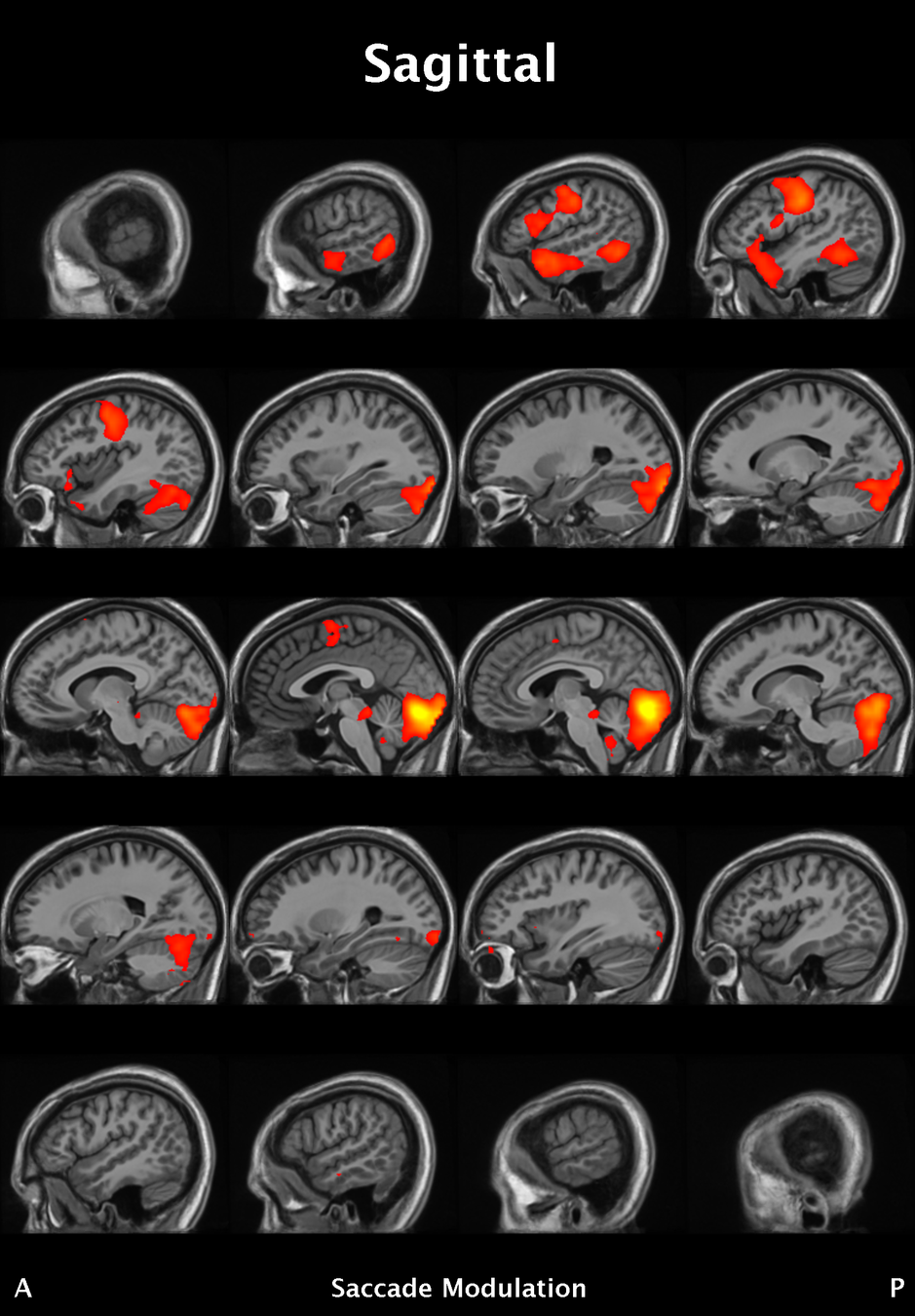
Where is this going?
Where is this going?
- Individual differences in reading measures are highly consistent over time in adults.
- Functional and structural comparisons.
- Begin to untangle language in the brain.
- Influence of word frequency, predictability and other lexical variables on HDR.
- Influence of linguistic factors on saccade-control related HDR as well as subcortical regions.
- Contrast healthy readers with unhealthy populations to determine diagnostic potential of reading.
Why Study This?
- An understanding of the neurological basis of reading is key to:
- Understand the physiology of language and reading related disorders.
- Develop/improve diagnostic methods for:
- Disorders of reading or language
- Oculomotor/Visual system
- Neuro/Psychopathology
- Improve therapies and track recovery or treatment efficacy.
- Improved quality of life.
- Inform strategies to teach others this vital skill.
Advisory Committee
- Dr. Steven Luke
- Dr. C. Brock Kirwan
- Dr. Derin Cobia
- Dr. Jonathan Wisco
- Dr. Michael Brown
BYU Eye-tracking Lab
- Jordan Olsen
- Brent Foster
- Trent Jackman
- Tanner Jensen
- Jason Lee
- Kristina Hall
- Steven Nish
BYU MRIF Associates and Staff


Questions?