An Introduction for Pythonic Data Scientists

Chad Scherrer
Senior Data Scientist, Metis

Follow Along Live:
What is Julia?
Language Features
- Built for performance from day 1
- JIT compilation
- Easy to call Python, R, C, Fortran
- Macros for automating code rewrites
- Multiple dispatch
Benefits
- Lowers user/dev barrier
- Less need to switch languages
- Better for pedagogy
People have assumed that we need both fast and slow languages. I happen to believe that we don't need slow languages.
- Jeff Bezanson, Julia co-creator
Plots.jl Ecosystem
Backends
PyPlot
Plotly / PlotlyJS
GR
UnicodePlots
PGFPlots
InspectDR
HDF5
StatsPlots
PlotRecipes
AtariAlgos
Reinforce
JuliaML
Augmentor
DifferentialEquations
PhyloTrees
EEG
ImplicitEquations
ControlSystems
ValueHistories
ApproxFun
AverageShiftedHistograms
MLPlots
LazySets
A Simple Plot

using Plots
using StatsPlots, Distributions
plot(Normal(1,2), legend=false, lw=2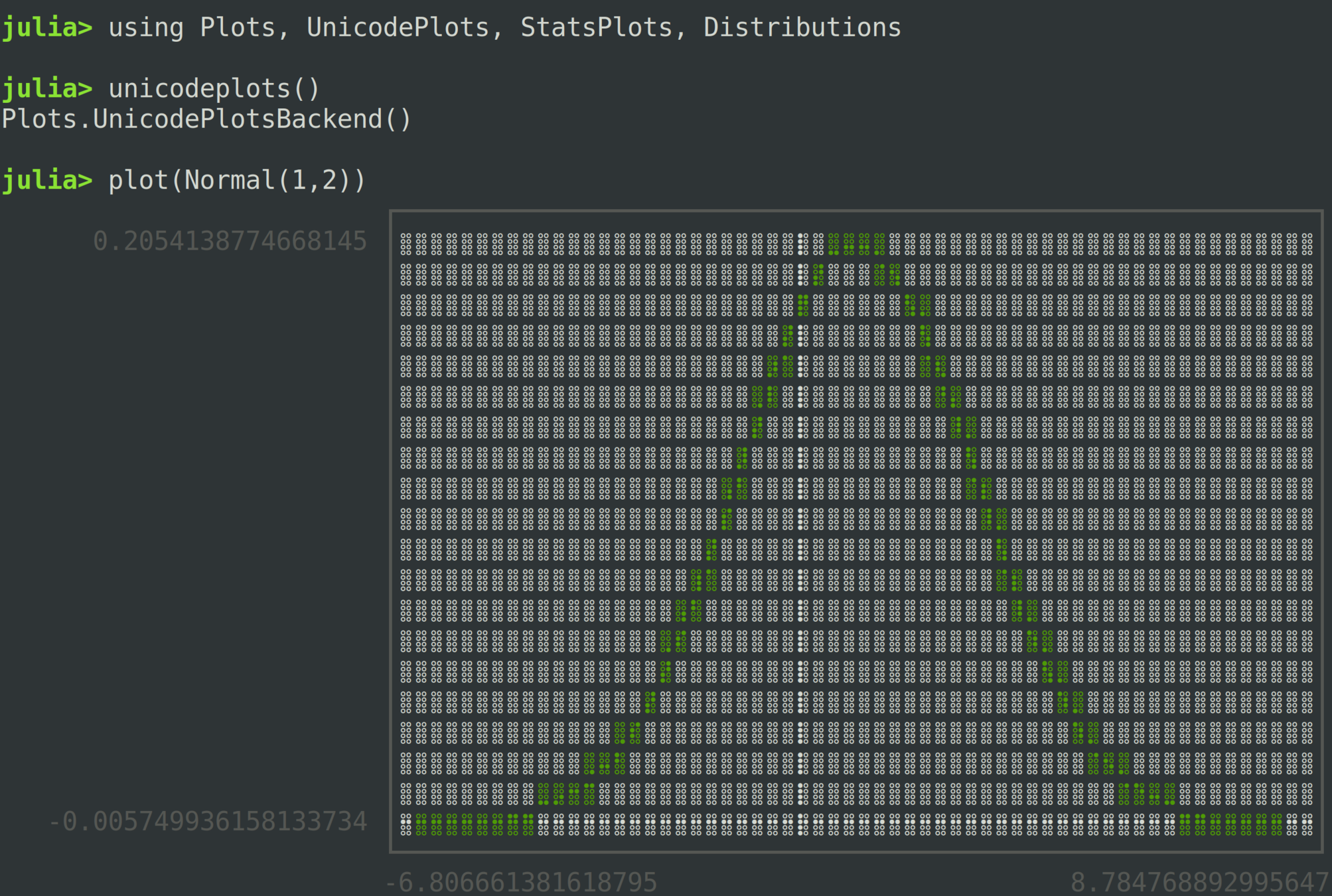
Easy GIFs!
plt = plot3d(1
, xlim=(-25,25)
, ylim=(-25,25)
, zlim=(0,50)
, title = "Lorenz Attractor"
, legend=false)
@gif for i=1:3000
step!(attractor)
push!(plt
, attractor.x
, attractor.y
, attractor.z)
end every 20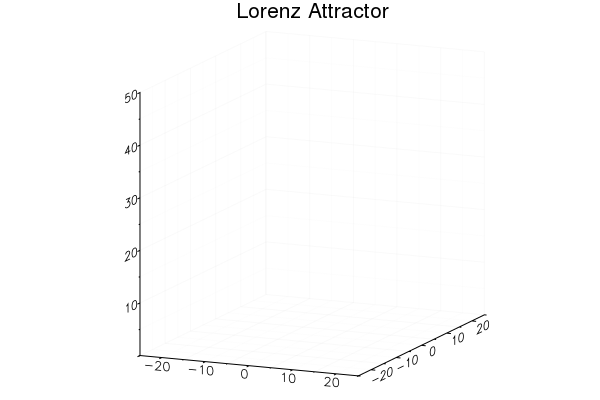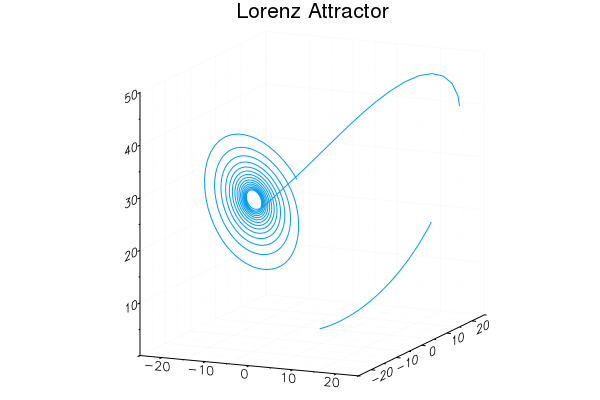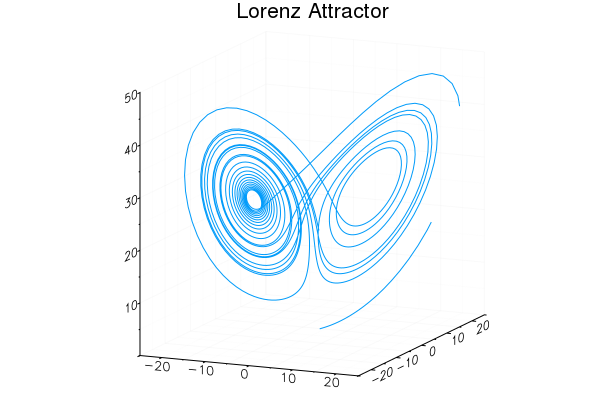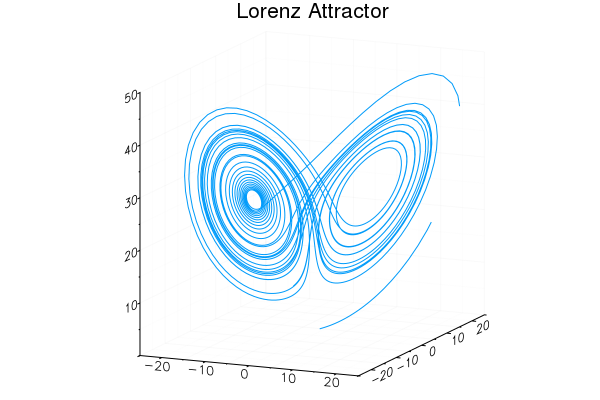
Side Quest
Using the Julia REPL
Measurements.jl
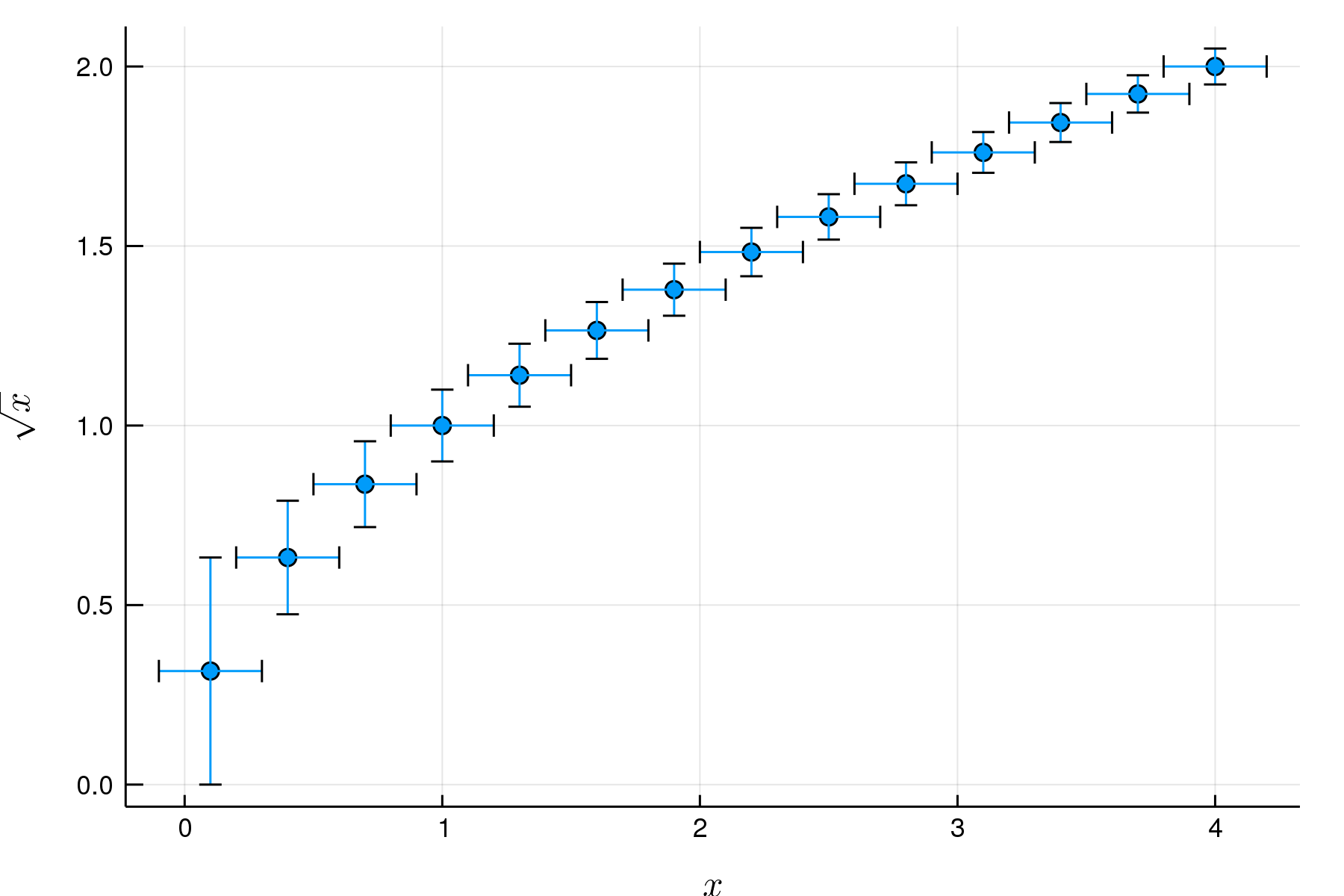
function mySqrt(x)
ans = (x + 1) / 2
for _ in 1:10
ans = (ans + x / ans) / 2
end
ans
end
x = (0.1 : 0.3 : 4) .± 0.2
y = mySqrt.(x)
scatter(x, y
, legend = false
, xlabel = L"x"
, ylabel = L"\sqrt x"
)ScikitLearn.jl
using GaussianMixtures: GMM
using ScikitLearn
gmm = fit!(GMM(n_components=3, kind=:diag), X)
predict_proba(gmm, X)- Interface to scikit-learn using PyCall.jl
- Easy access to Python methods
- All of scikit-learn, plus
- GaussianMixtures.jl
- DecisionTree.jl
- LowRankModels.jl
- Easy to add more!
TensorFlow.jl
i = tf.constant(0, name="i")
result = tf.constant(0, name="result")
output = tf.while_loop(
lambda i, result: tf.less(i, 10),
lambda i, result: [i+1, result+tf.pow(i,2)],
[i, result]
)@tf i = constant(0)
@tf result = constant(0)
output = @tf while i < 10
i_sq = i^2
[i=>i+1, result=>result+i_sq]
endPython
Julia
- Lots of advantages over the Python API
- Ready for TensorFlow 2.0
MXNet.jl
using MXNet
mlp = @mx.chain mx.Variable(:data) =>
mx.FullyConnected(name=:fc1, num_hidden=128) =>
mx.Activation(name=:relu1, act_type=:relu) =>
mx.FullyConnected(name=:fc2, num_hidden=64) =>
mx.Activation(name=:relu2, act_type=:relu) =>
mx.FullyConnected(name=:fc3, num_hidden=10) =>
mx.SoftmaxOutput(name=:softmax)
# data provider
batch_size = 100
include(Pkg.dir("MXNet", "examples", "mnist", "mnist-data.jl"))
train_provider, eval_provider = get_mnist_providers(batch_size)
# setup model
model = mx.FeedForward(mlp, context=mx.cpu())
# optimization algorithm
# where η is learning rate and μ is momentum
optimizer = mx.SGD(η=0.1, μ=0.9)
# fit parameters
mx.fit(model, optimizer, train_provider, n_epoch=20, eval_data=eval_provider)Flux.jl
encoder = Dense(28^2, N, leakyrelu) |> gpu
decoder = Dense(N, 28^2, leakyrelu) |> gpu
m = Chain(encoder, decoder)
loss(x) = mse(m(x), x)
evalcb = throttle(() -> @show(loss(data[1])), 5)
opt = ADAM()
@epochs 10 Flux.train!(
loss
, params(m)
, zip(data)
, opt
, cb = evalcb
)
m = Chain(
LSTM(N, 128),
LSTM(128, 128),
Dense(128, N),
softmax)
m = gpu(m)
function loss(xs, ys)
l = sum(crossentropy.(m.(gpu.(xs)), gpu.(ys)))
Flux.truncate!(m)
return l
end
opt = ADAM(0.01)
tx, ty = (Xs[5], Ys[5])
evalcb = () -> @show loss(tx, ty)
Flux.train!(loss, params(m), zip(Xs, Ys), opt,
cb = throttle(evalcb, 30))Autoencoder
Character RNN
Neural Differential Equations in Flux
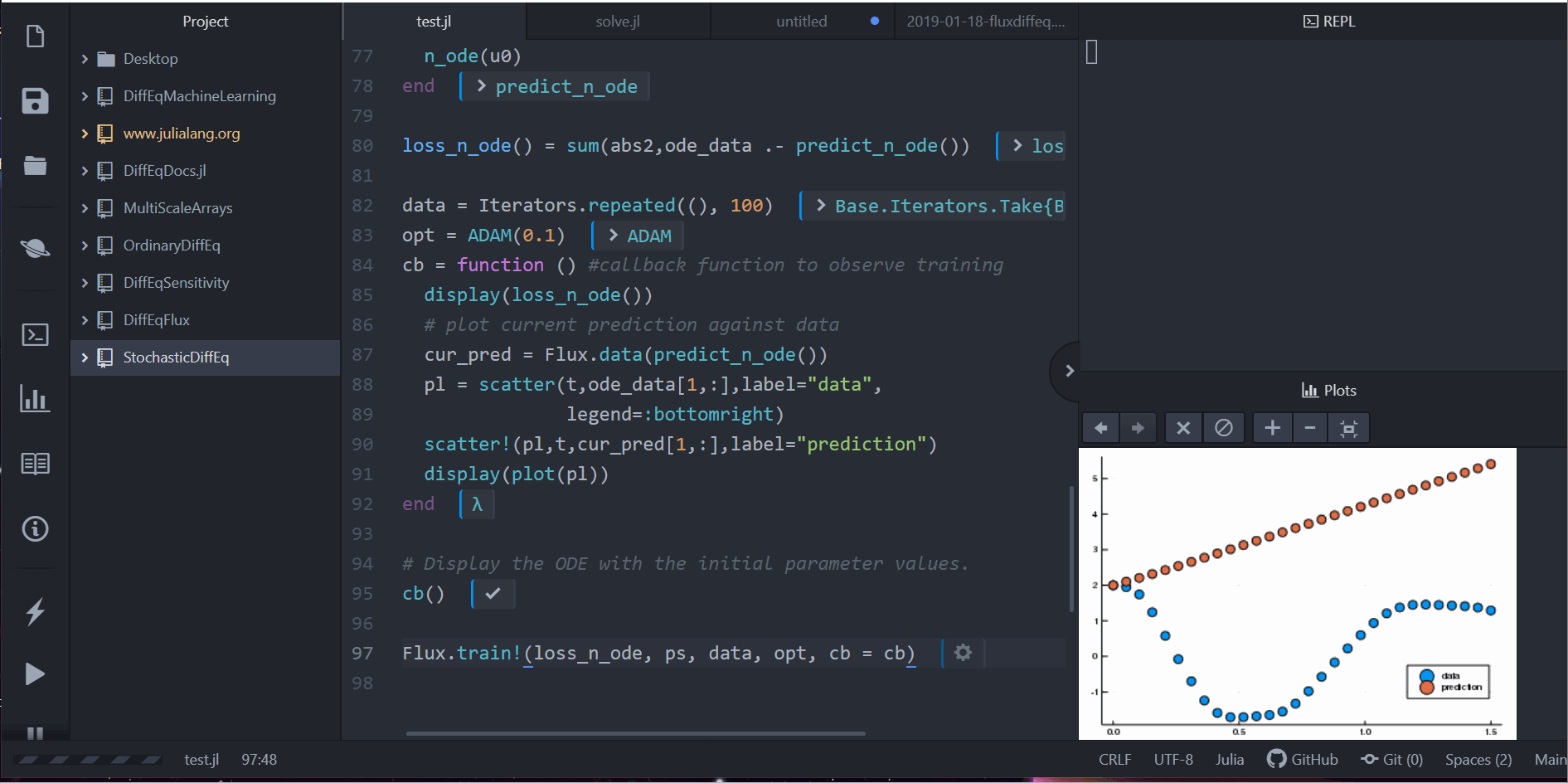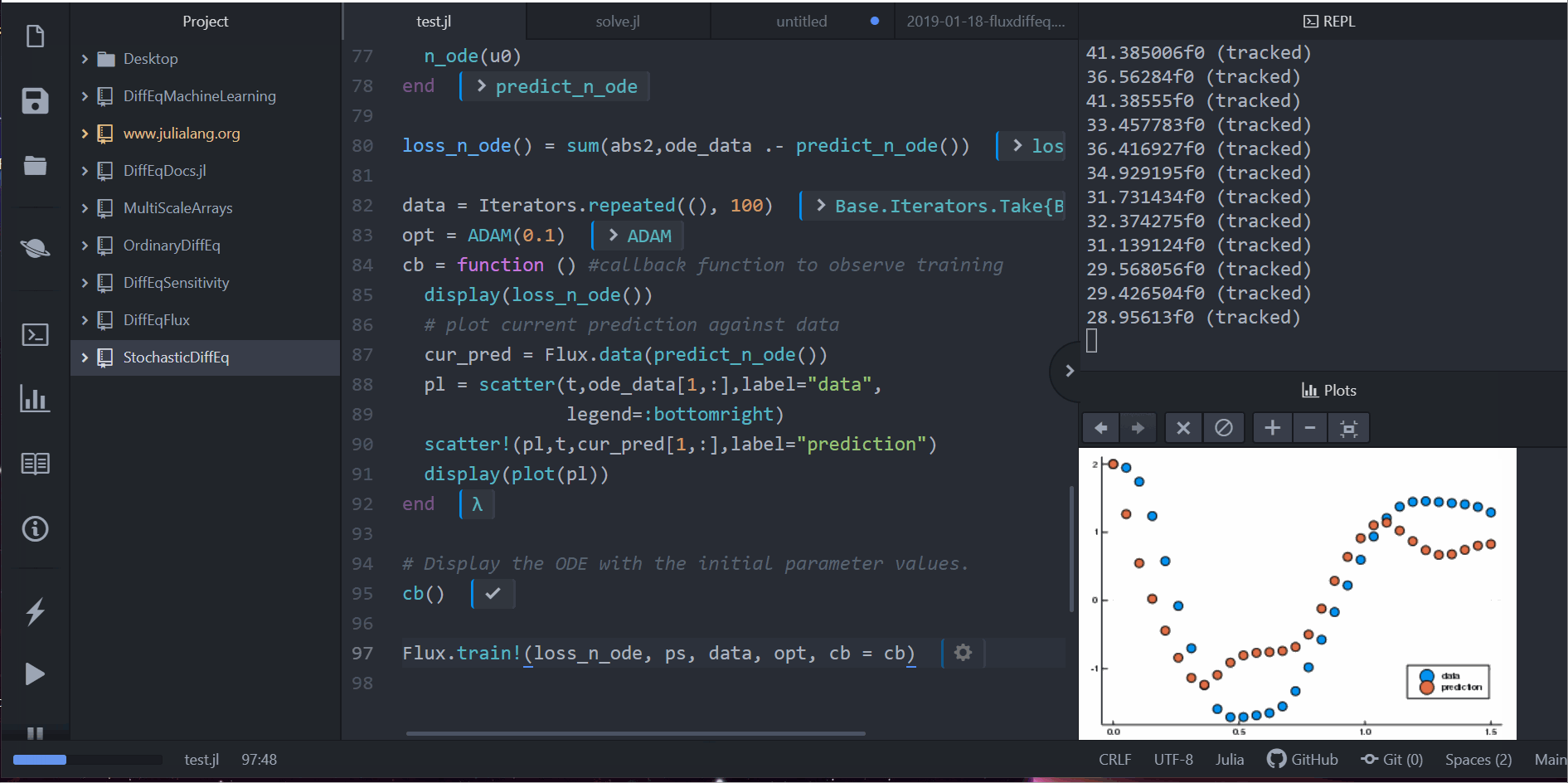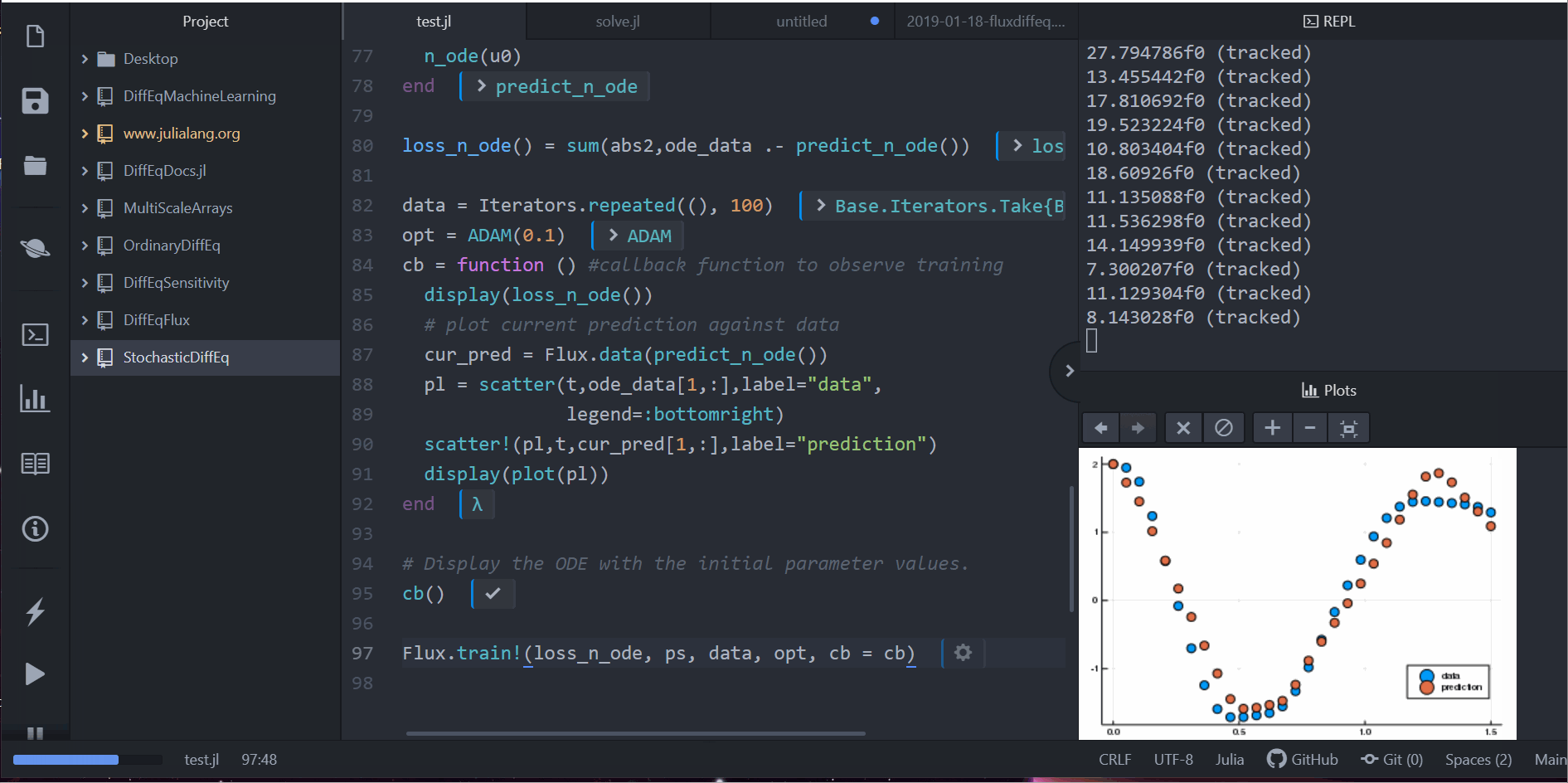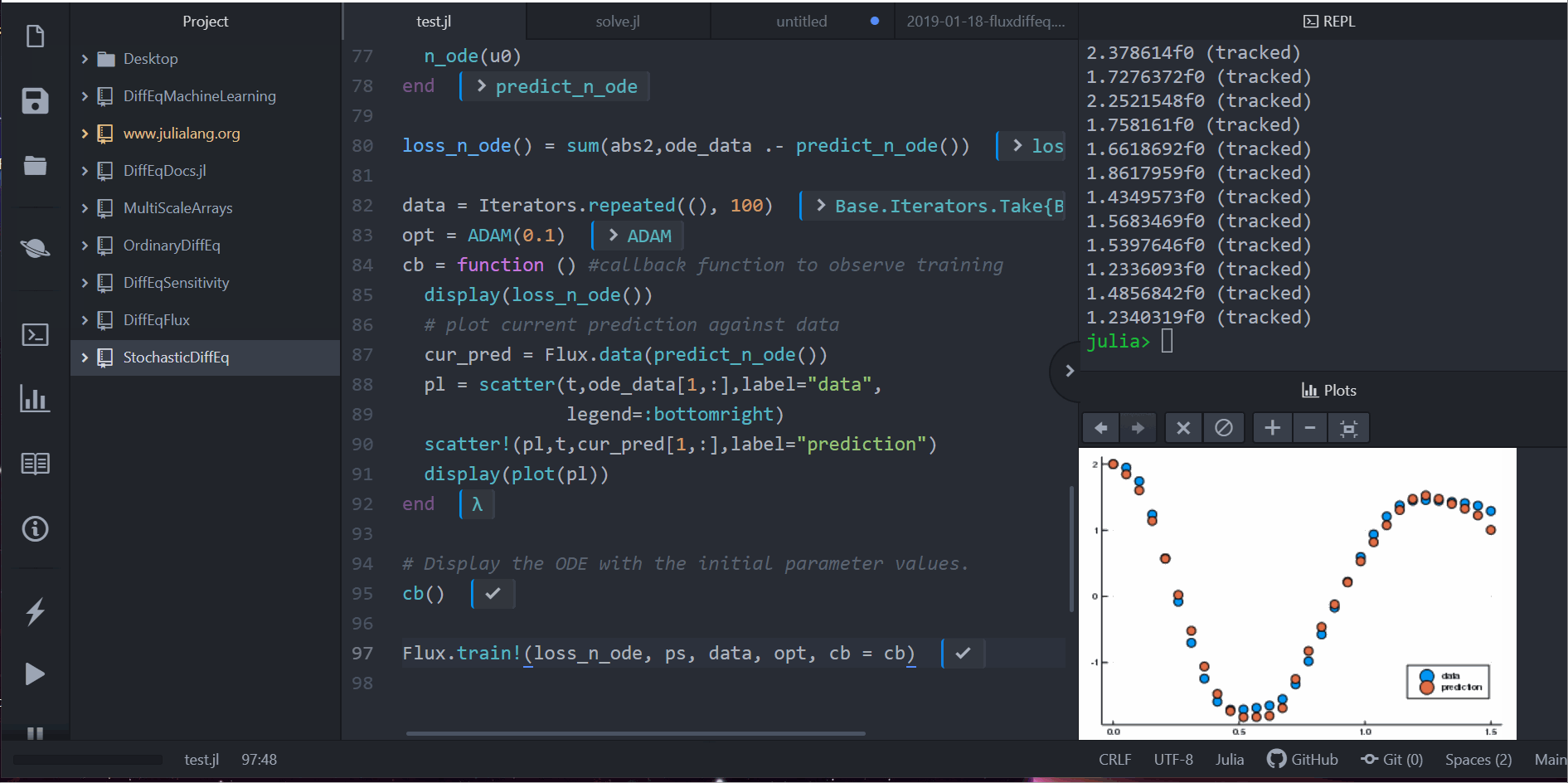
Lots More!
JuliaDB: Parallel out-of-core algorithms ("Dask with fast UDFs")
DataFrames: Pandas-like DataFrames
Queryverse: Common interface to query a wide variety of data sources
OnlineStats: O(n) algorithms
DifferentialEquations: Best-in-class DE algorithms
JuMP: Common interface for a wide variety of optimization libraries
Turing: Universal probabilistic programming
Soss: High-level Bayesian model rewriting (my project)
Cassette: Inject code transformations into JIT compilation cycle
Zygote: Source-to-source automatic differentiation
GPUArrays, CuArrays, CUDAnative
RCall: Call R from Julia
PyCall: Call Python from Julia
pyjulia: Call Julia from Python
Thank You!
