Neuroanatomical
Information
Systems
daniel.furth@ki.se
meletis lab
karolinska institutet, SWEDEN
Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.
Sidney Brenner
-
A web-based and open source stereotactic mouse brain atlas.
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
>800'000 neurons in 21 brains.
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
>800'000 neurons in 21 brains.
-
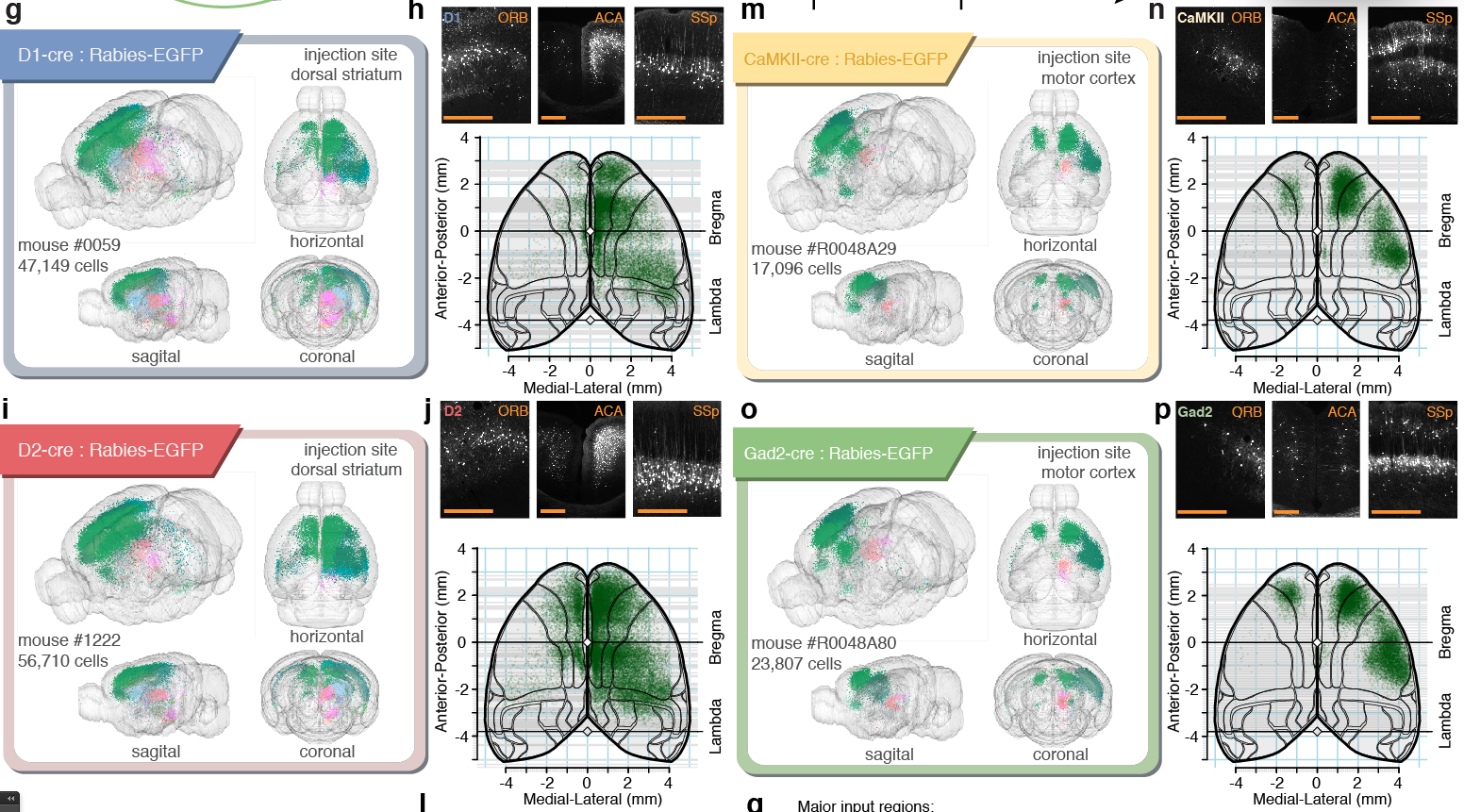
Cell-type specific connectivity tracing in five different Cre-lines (D1, D2, ChAT, Camk2, Gad2).
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
>800'000 neurons in 21 brains.
-
Cell-type specific connectivity tracing in five different Cre-lines (D1, D2, ChAT, Camk2, Gad2).
-
Mapping out connectivity of ~160'000 neurons in the corticostriatal pathway (~0.23% of the entire mouse brain).
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
>800'000 neurons in 21 brains.
-
Cell-type specific connectivity tracing in five different Cre-lines (D1, D2, ChAT, Camk2, Gad2).
-
Mapping out connectivity of ~160'000 neurons in the corticostriatal pathway (~0.23% of the entire mouse brain).
-
Simultaneous segmentation of cell bodies and fiber tracts.
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
>800'000 neurons in 21 brains.
-
Cell-type specific connectivity tracing in five different Cre-lines (D1, D2, ChAT, Camk2, Gad2).
-
Mapping out connectivity of ~160'000 neurons in the corticostriatal pathway (~0.23% of the entire mouse brain).
-
Simultaneous segmentation of cell bodies and fiber tracts.
-
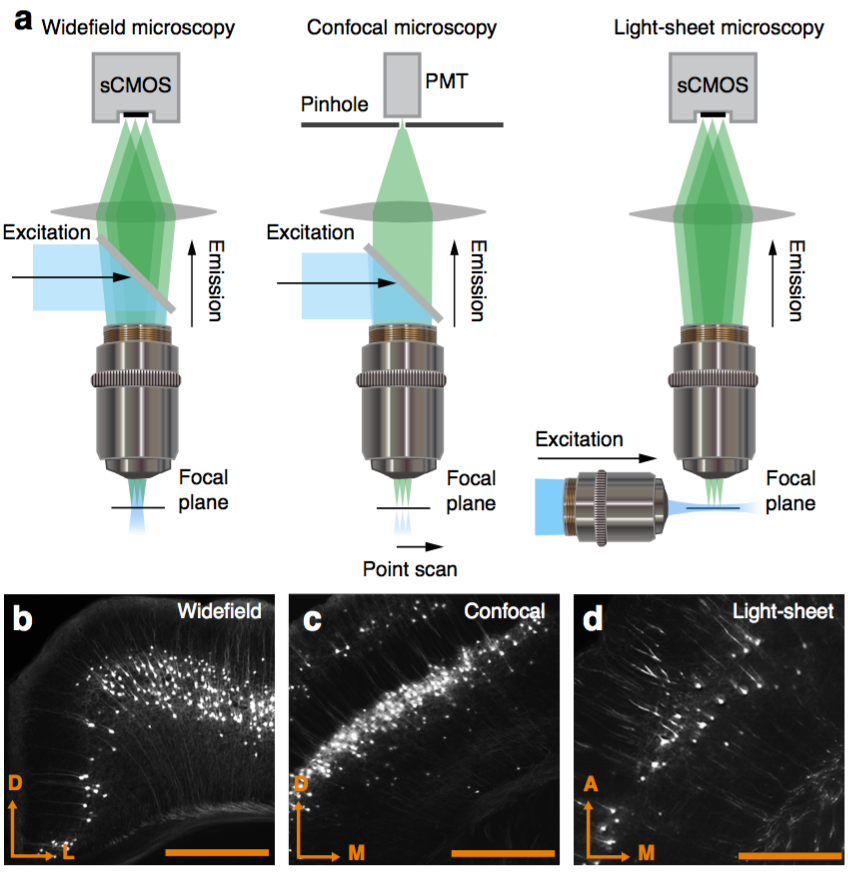
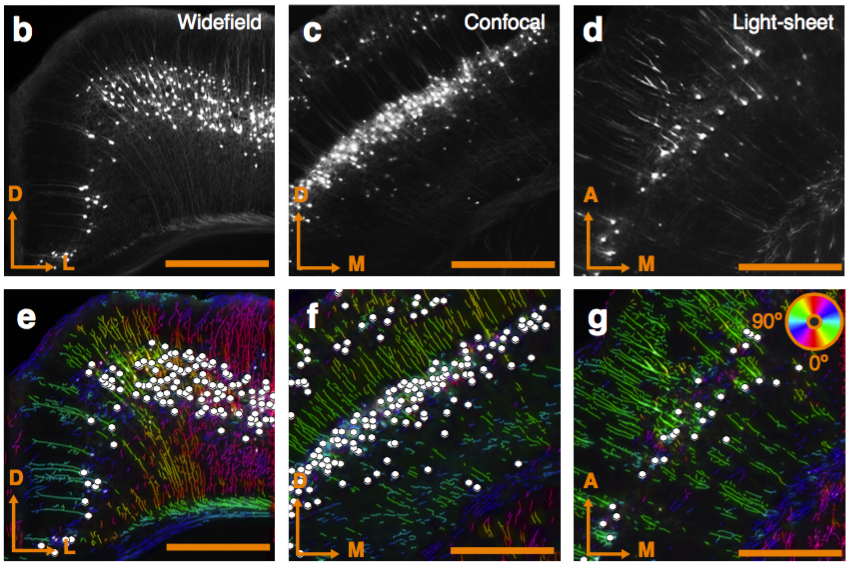
Works with FP, IHC, ISH, widefield, confocal, light-sheet.
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
>800'000 neurons in 21 brains.
-
Cell-type specific connectivity tracing in five different Cre-lines (D1, D2, ChAT, Camk2, Gad2).
-
Mapping out connectivity of ~160'000 neurons in the corticostriatal pathway (~0.23% of the entire mouse brain).
-
Simultaneous segmentation of cell bodies and fiber tracts.
-
Works with FP, IHC, ISH, widefield, confocal, light-sheet.
-
Reconstructing an entire brain in 20 min.
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
>800'000 neurons in 21 brains.
-
Cell-type specific connectivity tracing in five different Cre-lines (D1, D2, ChAT, Camk2, Gad2).
-
Mapping out connectivity of ~160'000 neurons in the corticostriatal pathway (~0.23% of the entire mouse brain).
-
Simultaneous segmentation of cell bodies and fiber tracts.
-
Works with FP, IHC, ISH, widefield, confocal, light-sheet.
-
Reconstructing an entire brain in 20 min.
-
Compression of large TIFF files (>300 Mb) to ~20 Mb.
-
A web-based and open source stereotactic mouse brain atlas.
-
Ability to annotate and query.
-
>800'000 neurons in 21 brains.
-
Cell-type specific connectivity tracing in five different Cre-lines (D1, D2, ChAT, Camk2, Gad2).
-
Mapping out connectivity of ~160'000 neurons in the corticostriatal pathway (~0.23% of the entire mouse brain).
-
Simultaneous segmentation of cell bodies and fiber tracts.
-
Works with FP, IHC, ISH.
-
Reconstructing an entire brain in 20 min.
-
Compression of large TIFF files (>300 Mb) to ~20 Mb.
-
Sharing data and images easy over the web.
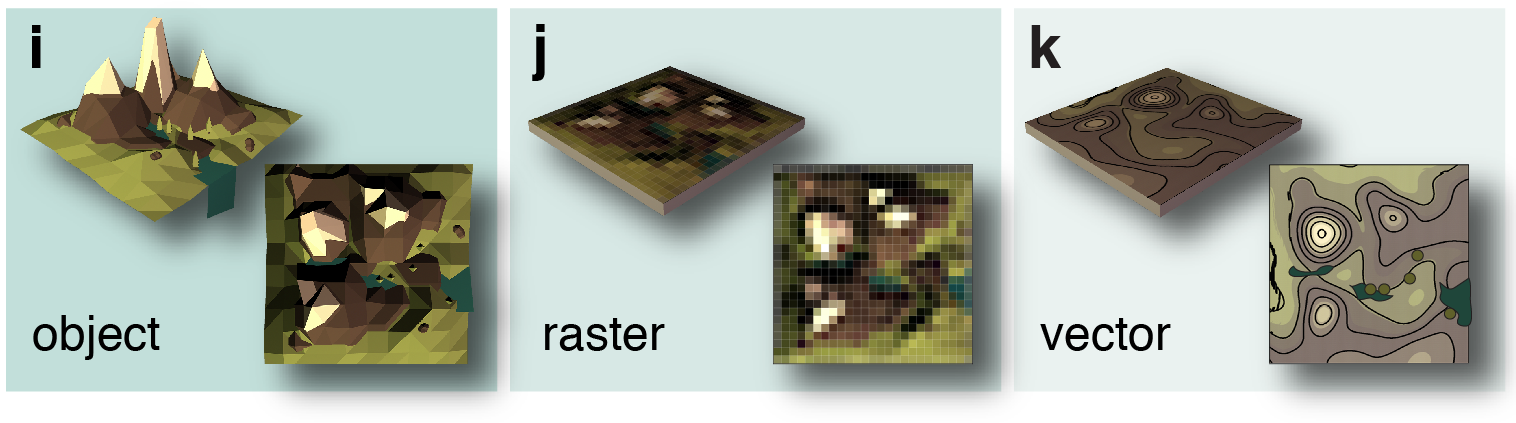
Geographical Information Systems (GIS)
Geographical Information Systems (GIS)
Raster graphics in neuroscience
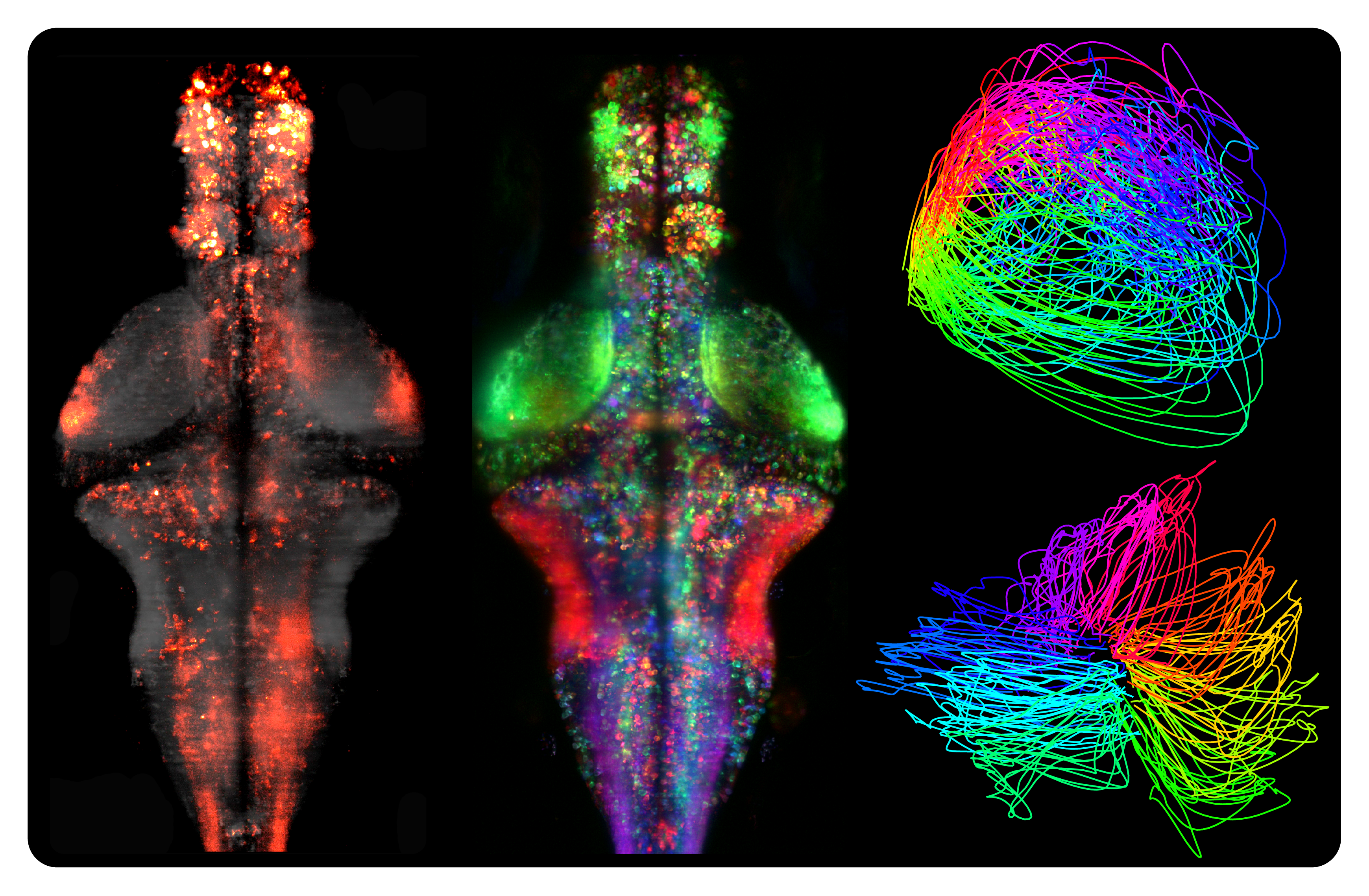
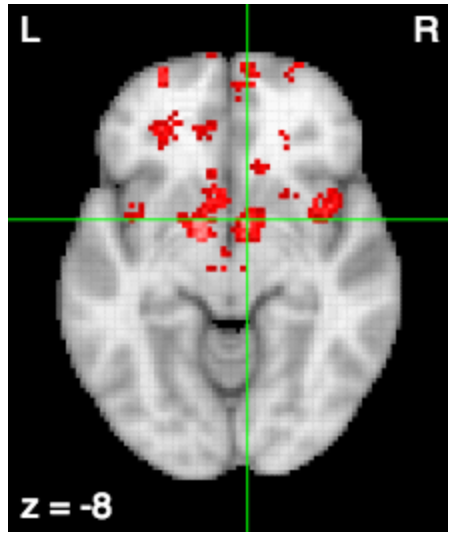
Freeman et al. (2014) Nature Methods
Raster graphics in neuroscience

8204 citations
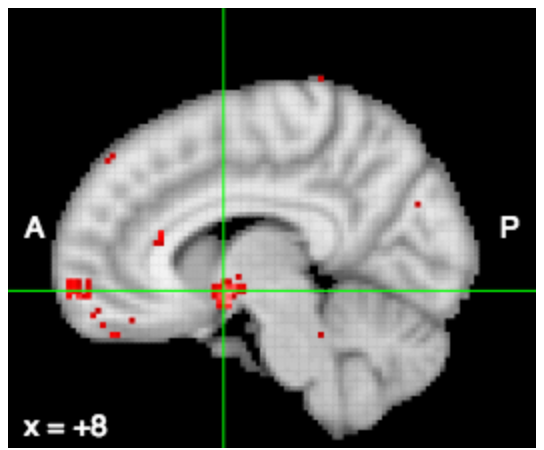
Friston et al. (1994) Human Brain Mapping
Raster graphics in neuroscience

8204 citations
Friston et al. (1994) Human Brain Mapping
Raster graphics in neuroscience

Raster graphics in neuroscience

Raster graphics in neuroscience

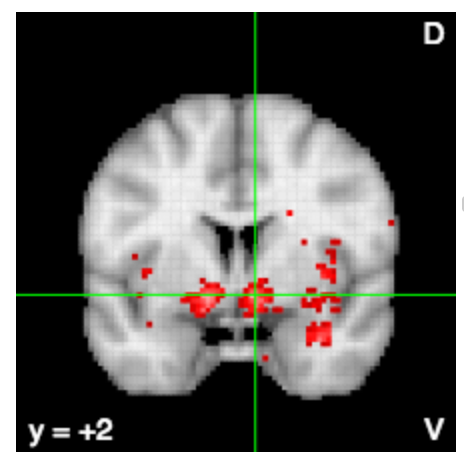
food, weight, obese, eating, foods, obesity, reward, energy, hunger, calorie, intake, bmi, mass, body, satiety, normal, cues, responsivity, women, overweight, appetite, individuals, caloric, increases, lean, index, fat, pictures, gain, hungry, images, bed, palatable, craving, receipt, water, overeating, meal, kg, motivation
neurosynth.org
Vector graphics in neuroscience
?
Vector graphics in neuroscience
Bakker, Tiesinga, Kötter (2015) Neuroinformatics
Why does it matter?
Raster graphics:
Vector graphics:
Neuroanatomical Information Systems (NIS)

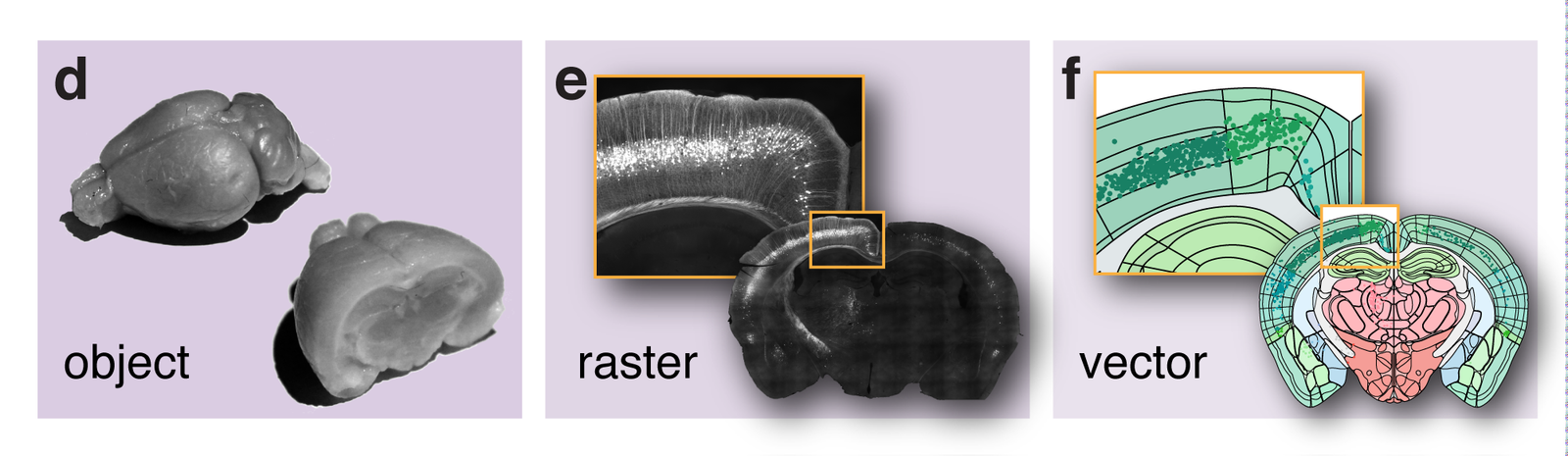
Neuroanatomical Information Systems (NIS)
Image processing tools in biology

-
ITK
- The Insight Segmentation and Registration Toolkit
- Visible Human Project (1986-1995)
- U.S. National Library of Medicine (NLM)
- Joseph Paul Jernigan (1954-1993)
-
VTK
- Visualization Toolkit
- GE Medical Systems
-
Sandia National Laboratories (Dept. Energy)
- KitWare
ParaView
-
ImageJ
- NIH Image (Object Pascal, macintosh only)
- ImageJ (1997)
- Fiji/Imagej (2009)
Pavel Tomancak







_Logo.svg/2000px-FIJI_(software)_Logo.svg.png)
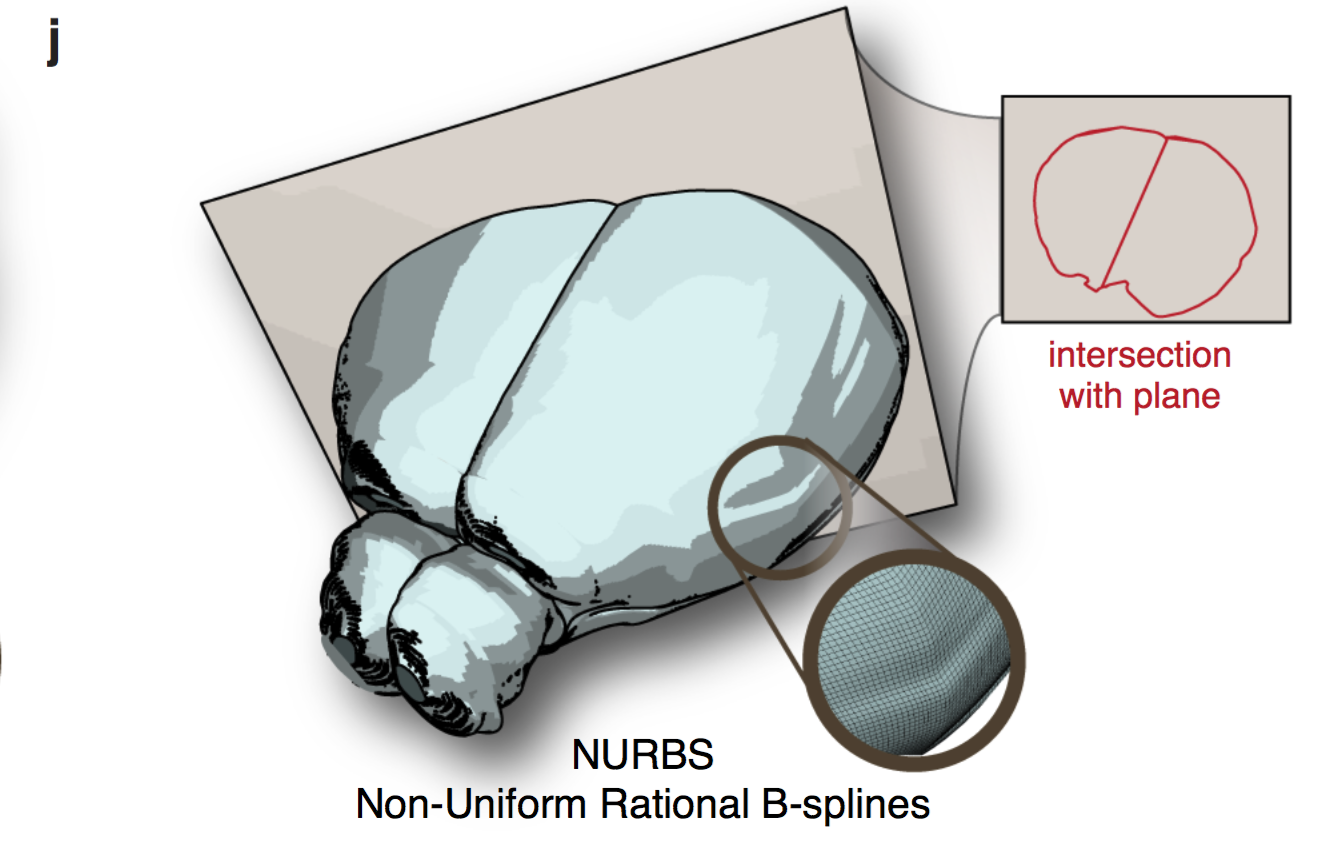
Scale-invariant atlas
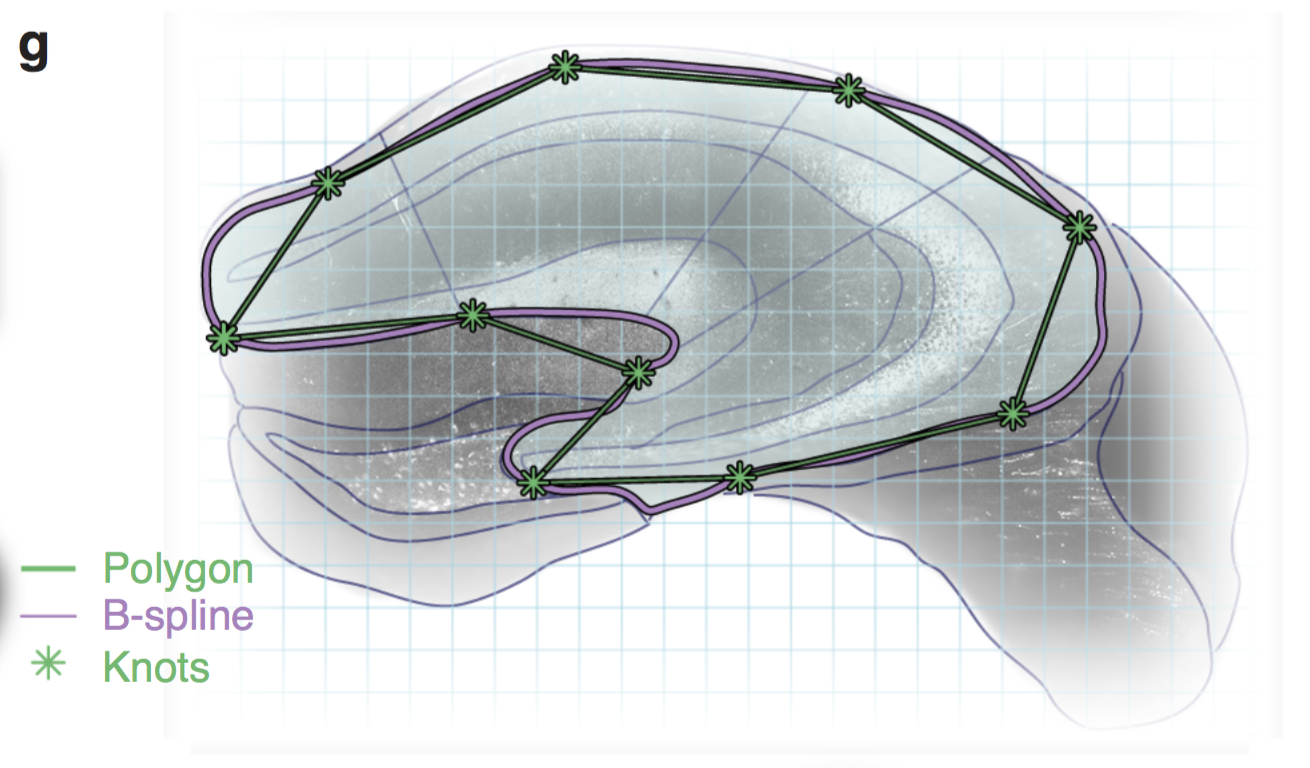
Scale-invariant atlas
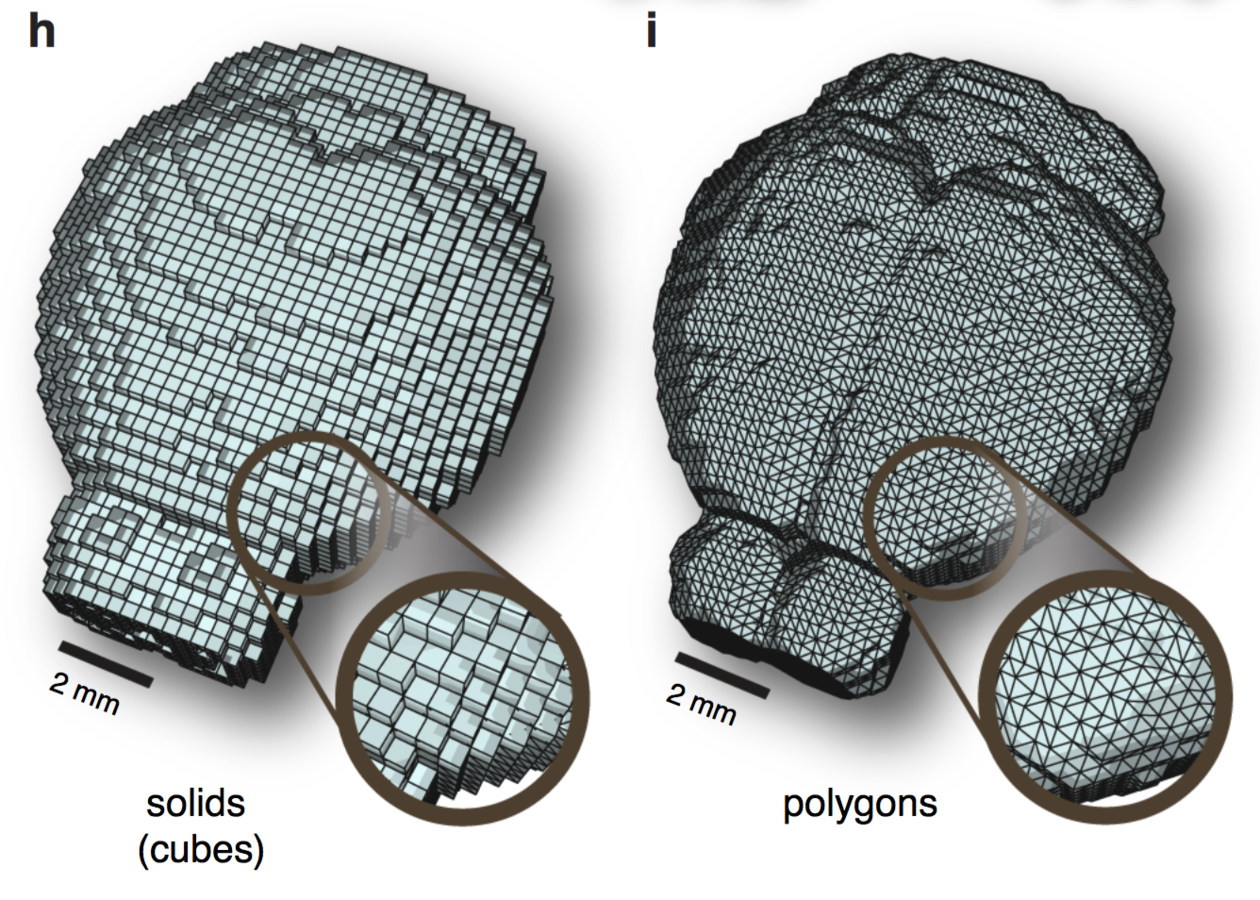
Scale-invariant atlas
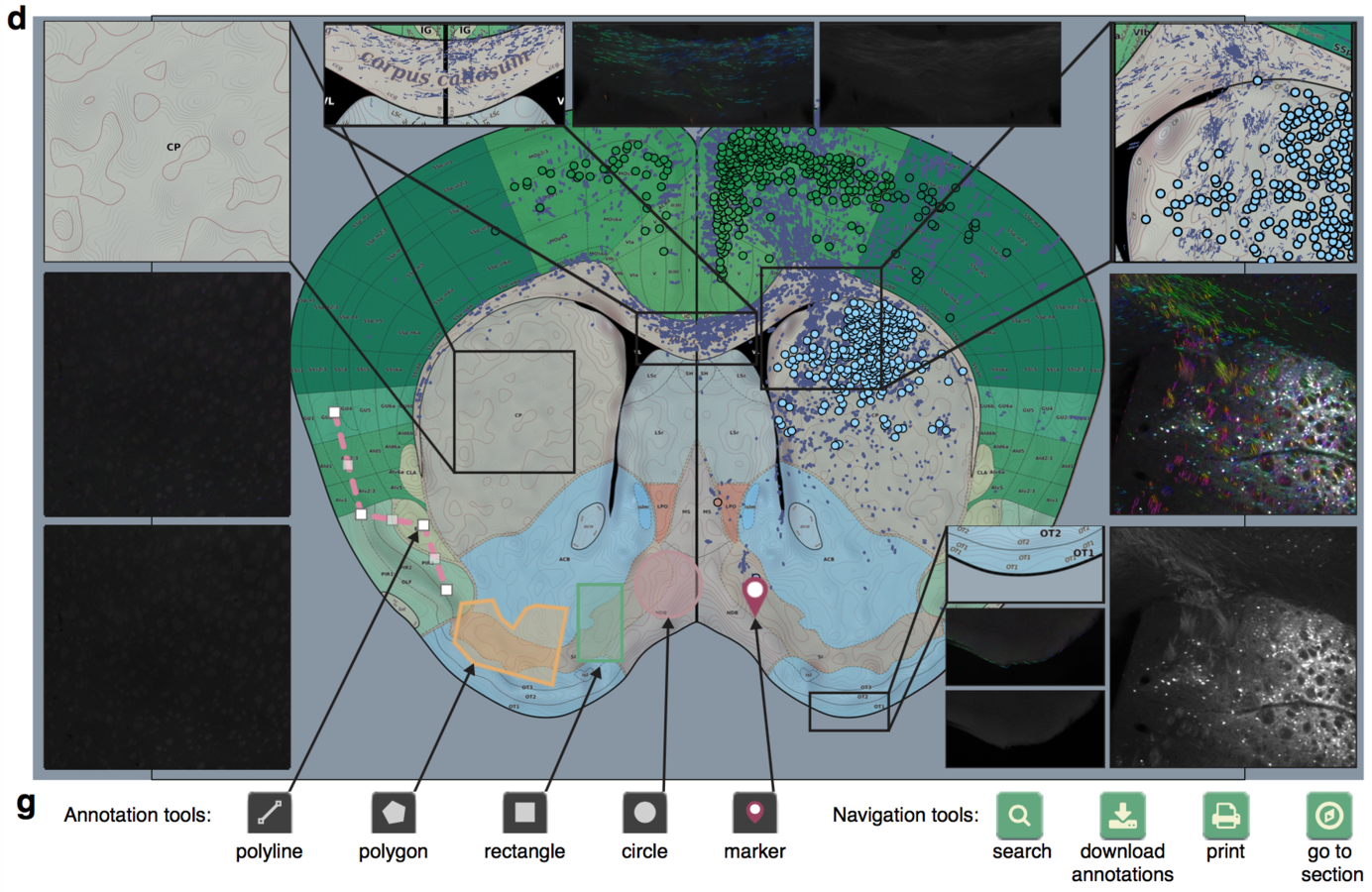
Web based interface
Web based interface
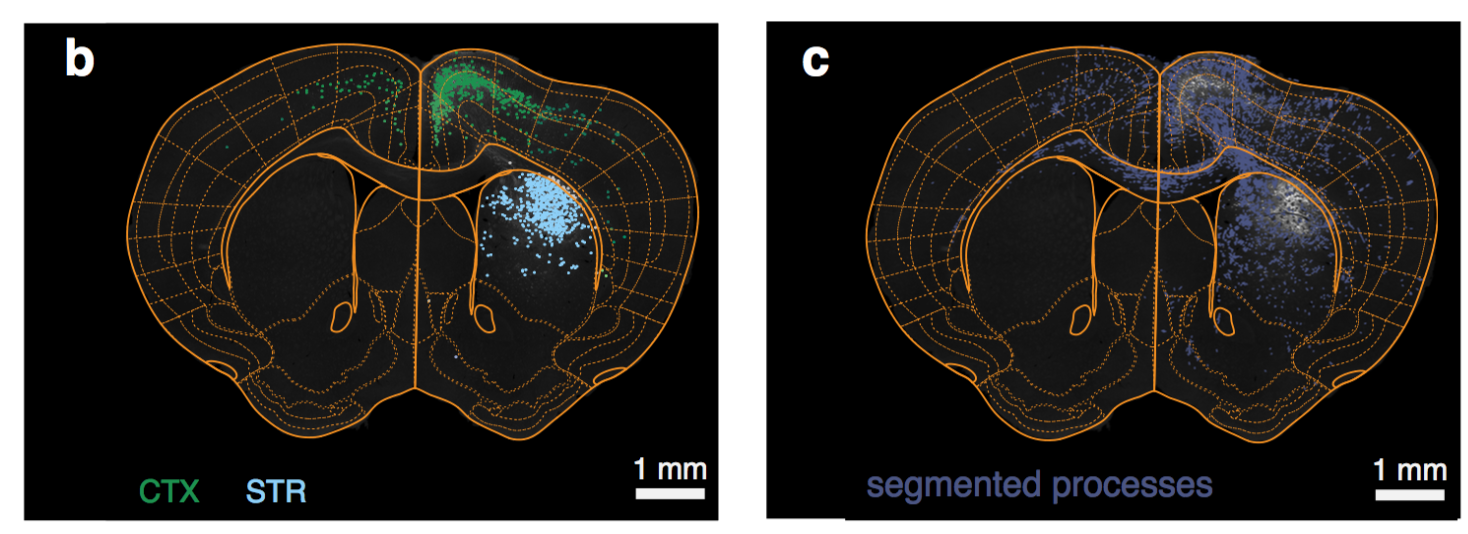
Web based interface
Web based interface
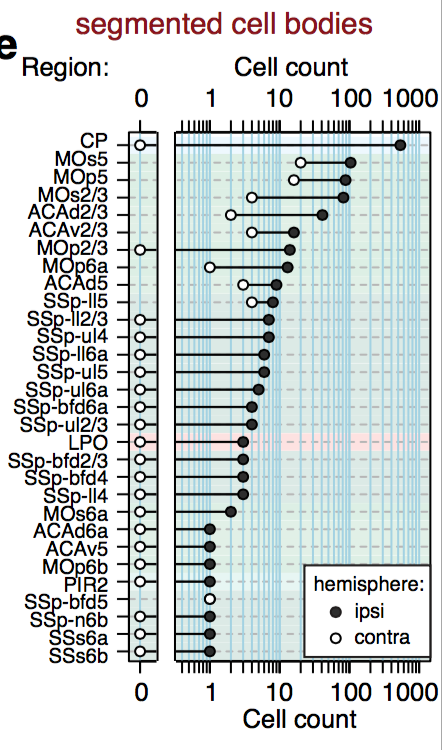
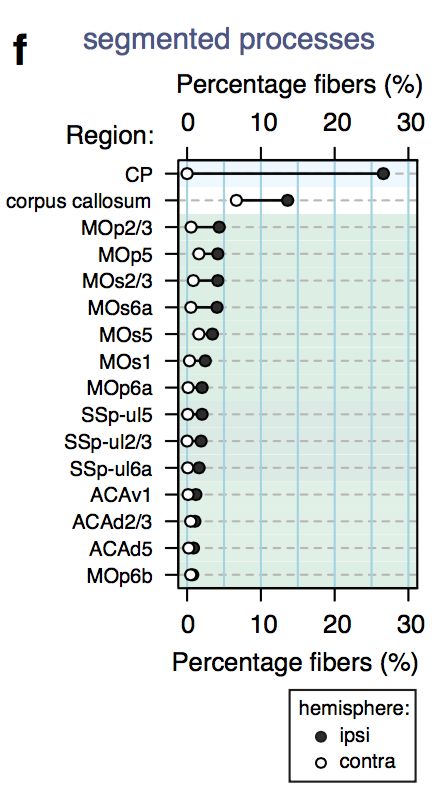
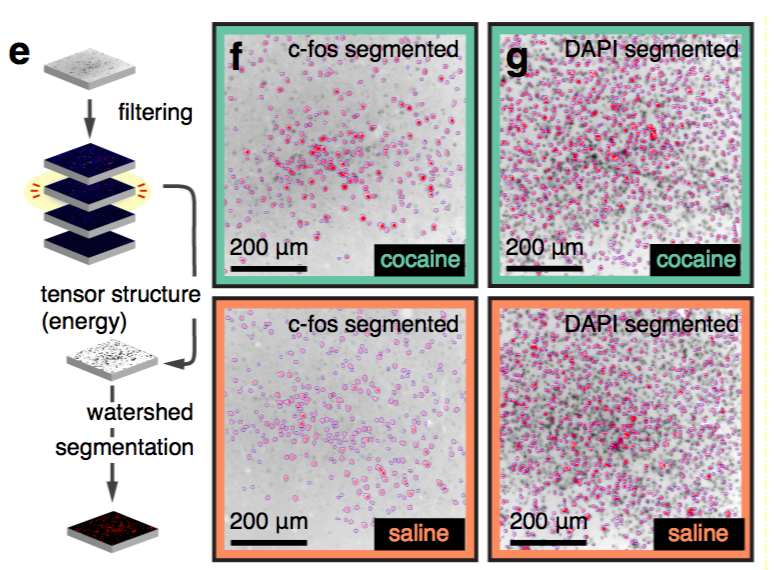
Segmentation
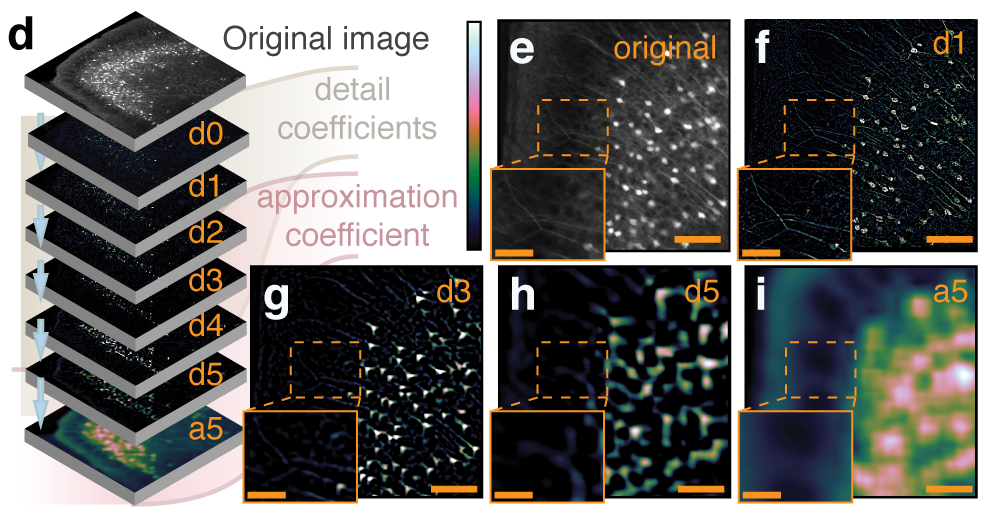
Segmentation
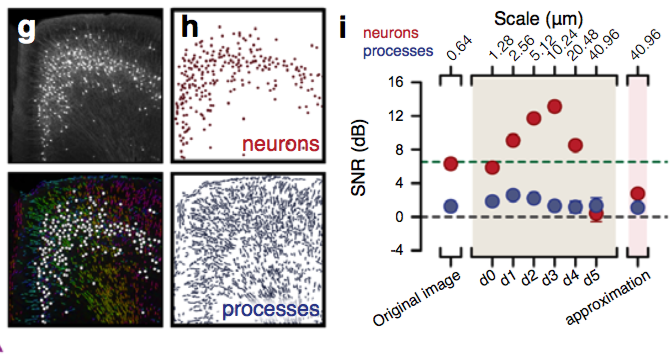
Segmentation
Segmentation
Segmentation
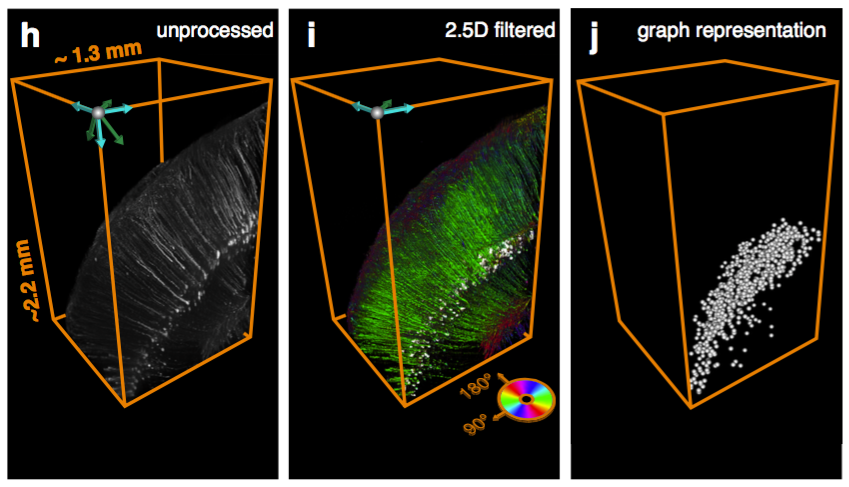
Registration
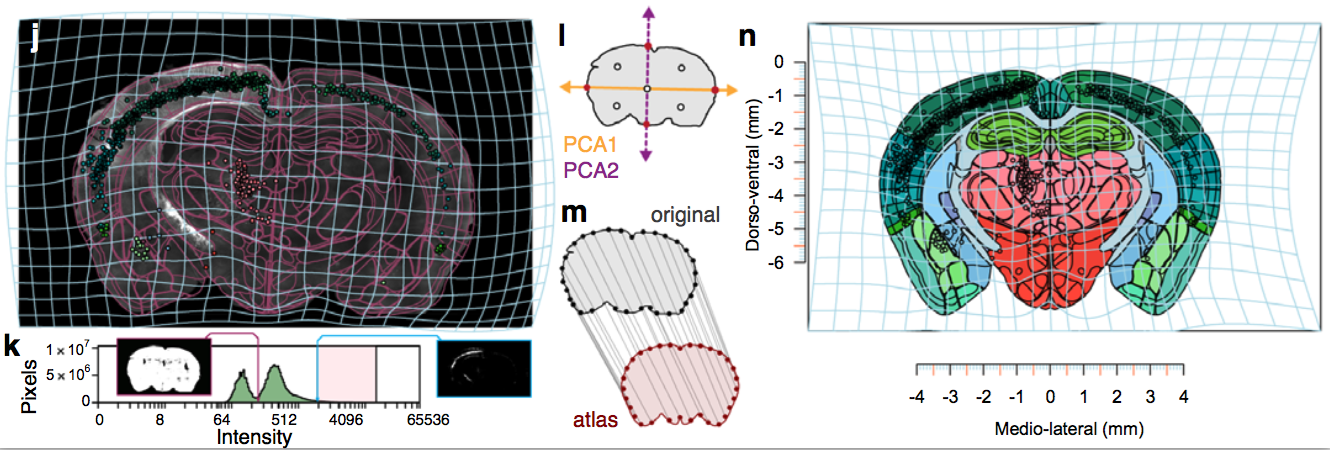
Segmentation and registration
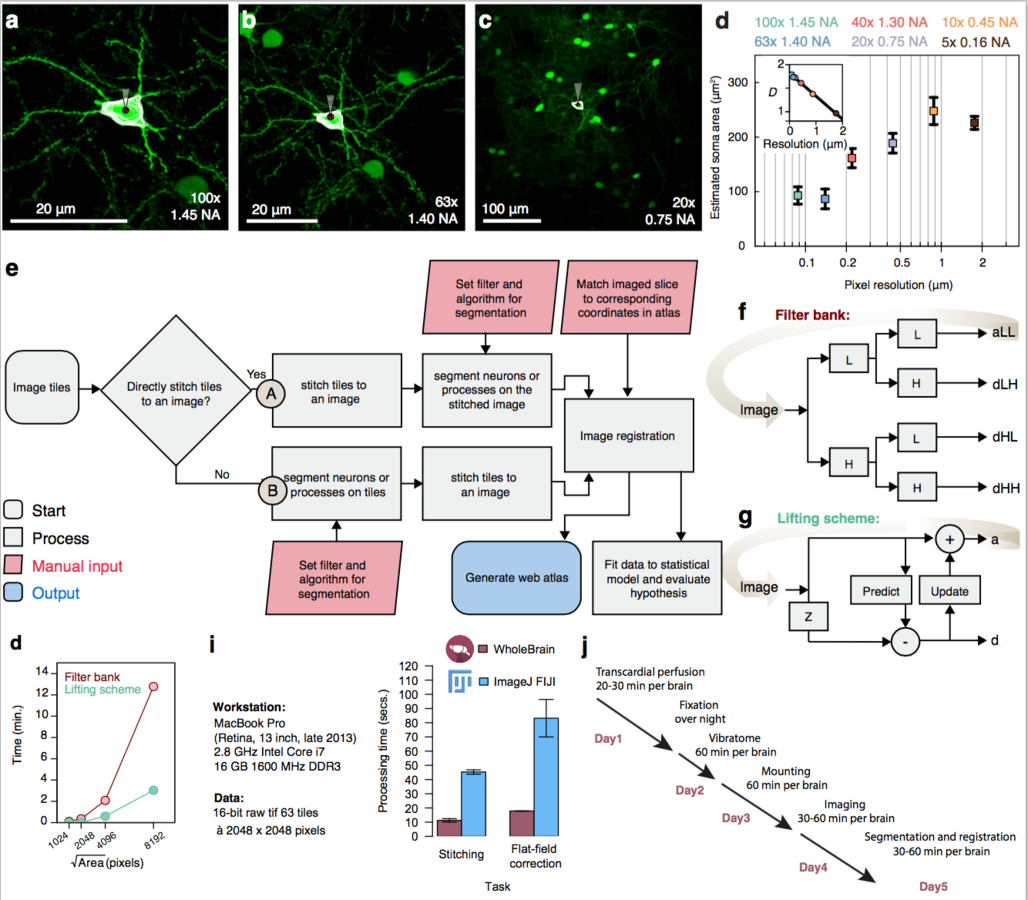
Segmentation and registration
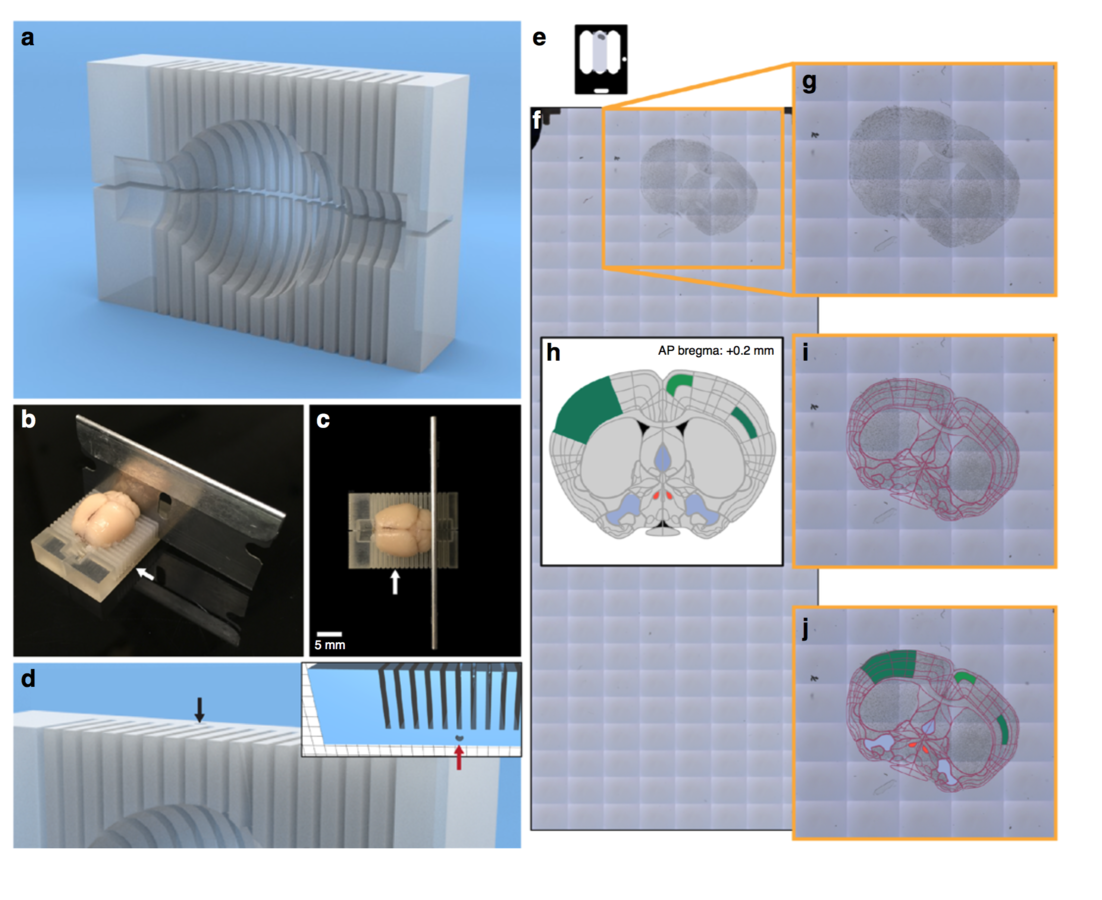
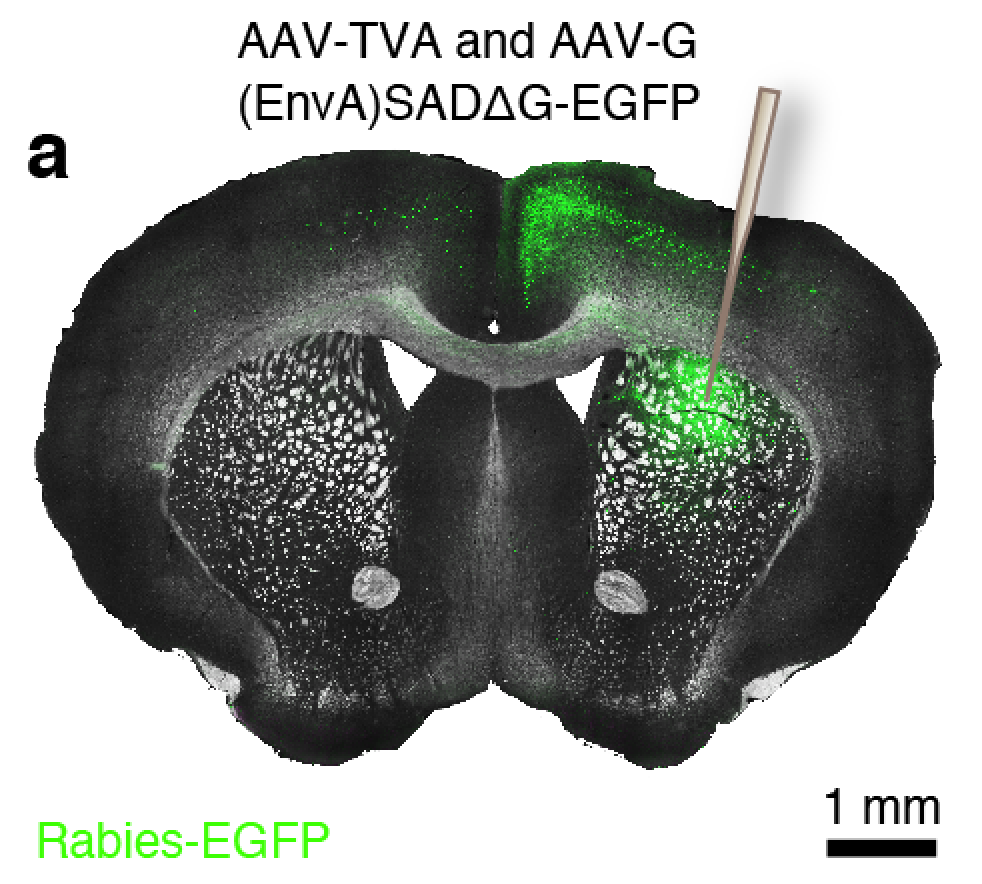
Connectivity tracing
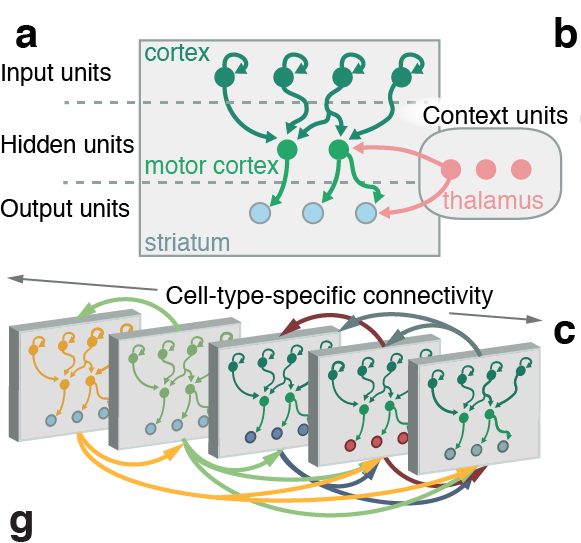
Connectivity tracing
Connectivity tracing
Connectivity tracing
Connectivity tracing
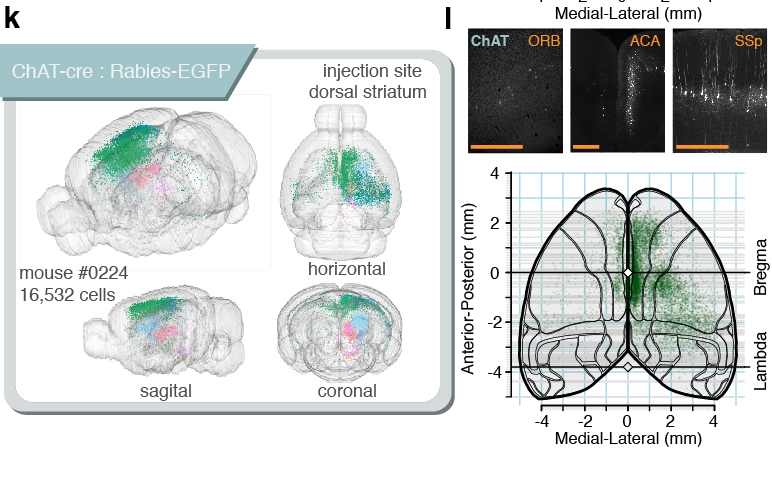
Connectivity tracing
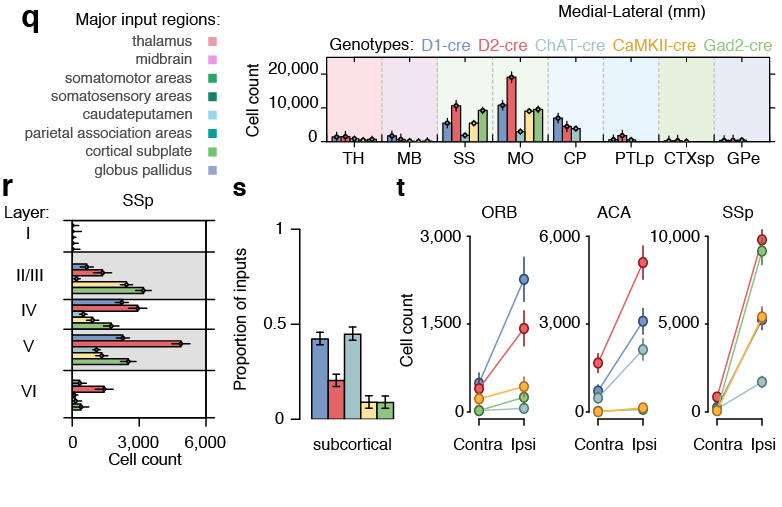
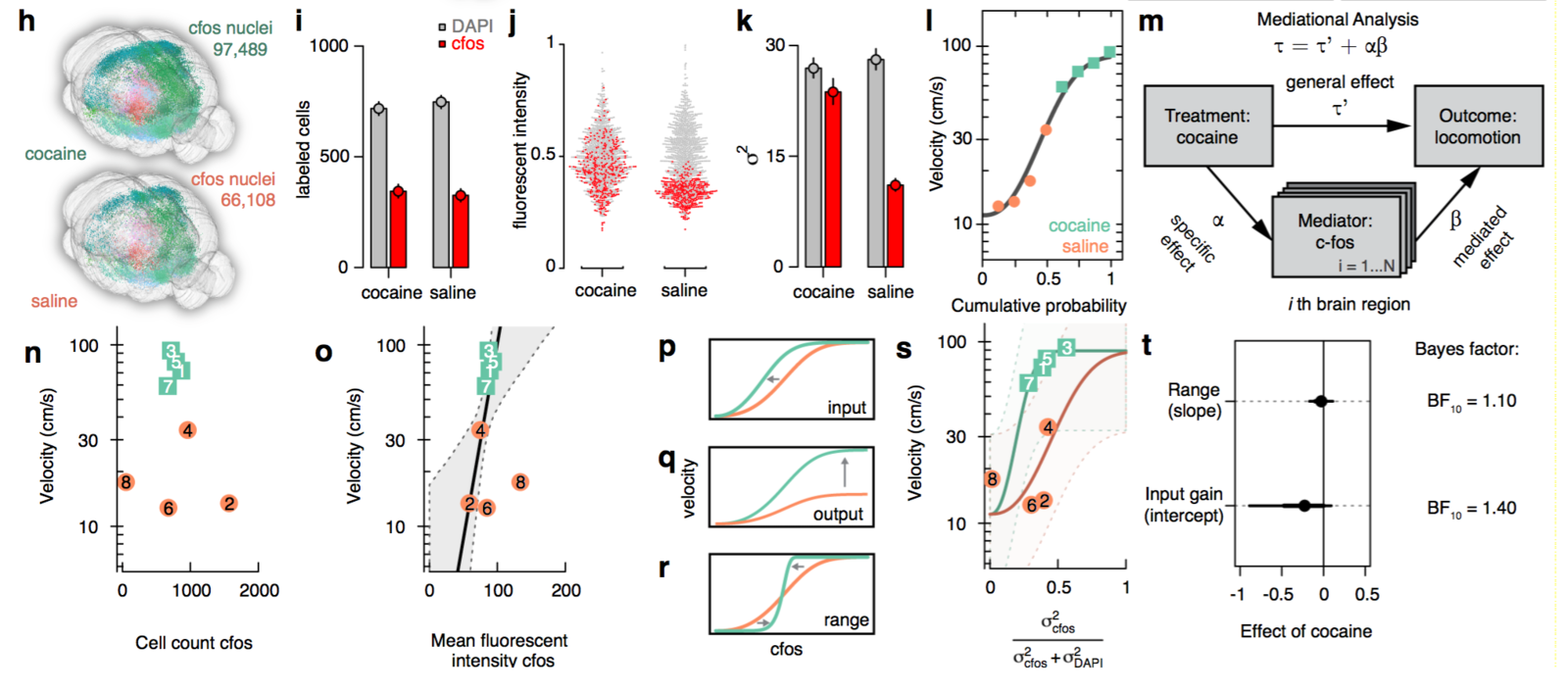
Decoding motor activity
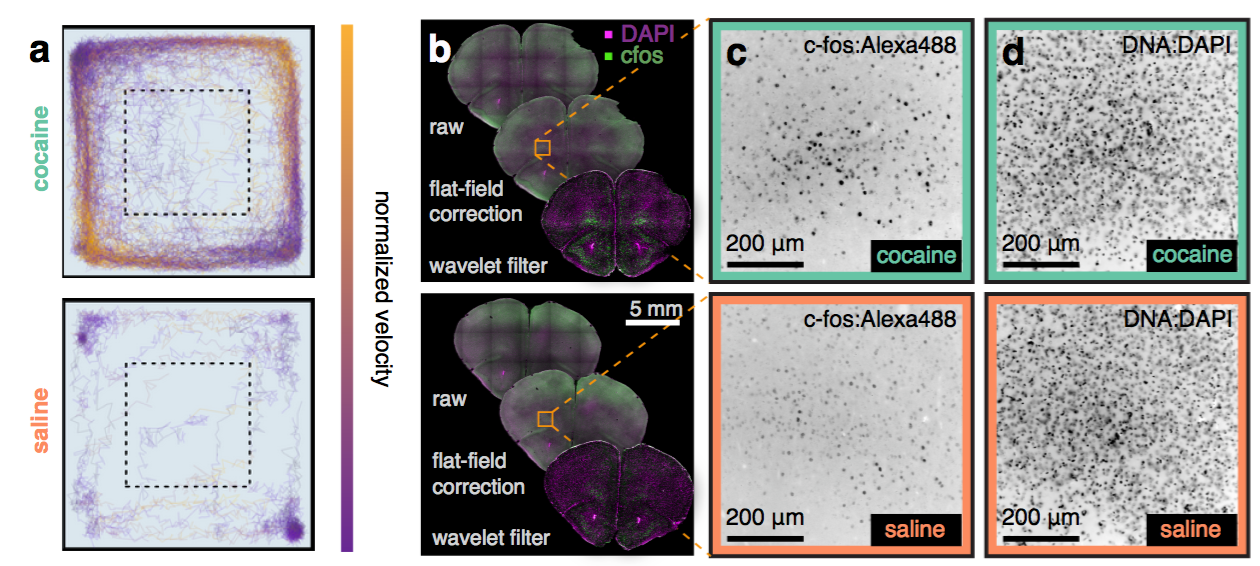
Decoding motor activity
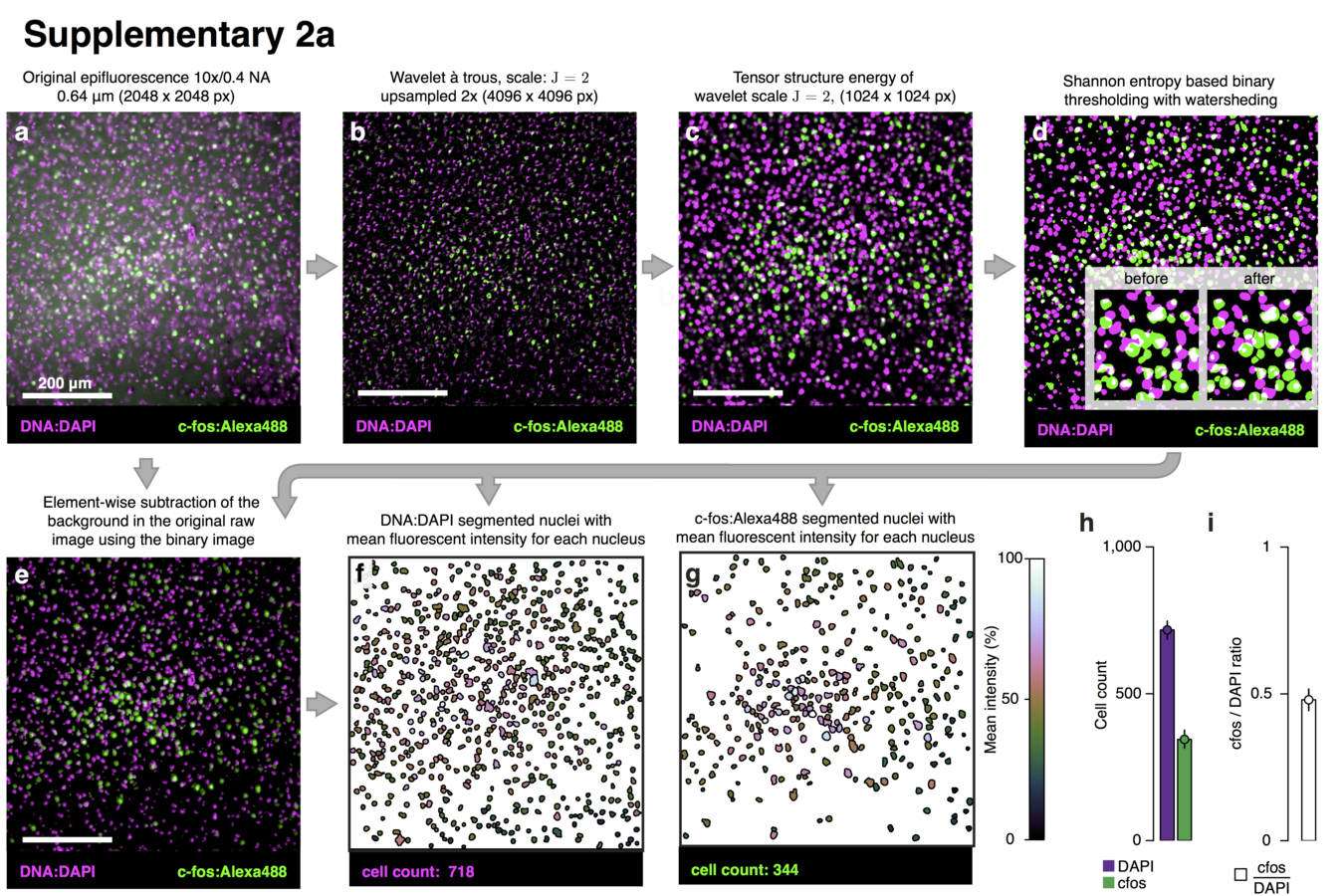
Decoding motor activity
Decoding motor activity
Barcoding connections
Barcoding connections
What is needed?
A method that:
- uniquely labels single postsynaptic neurons
- uniquely labels their presynaptic partners
- uniquely label axons and trace them
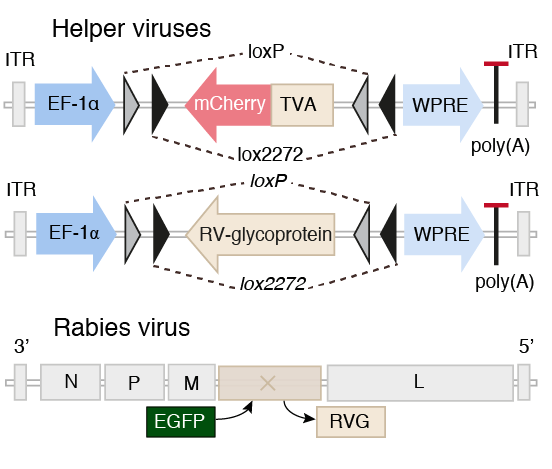
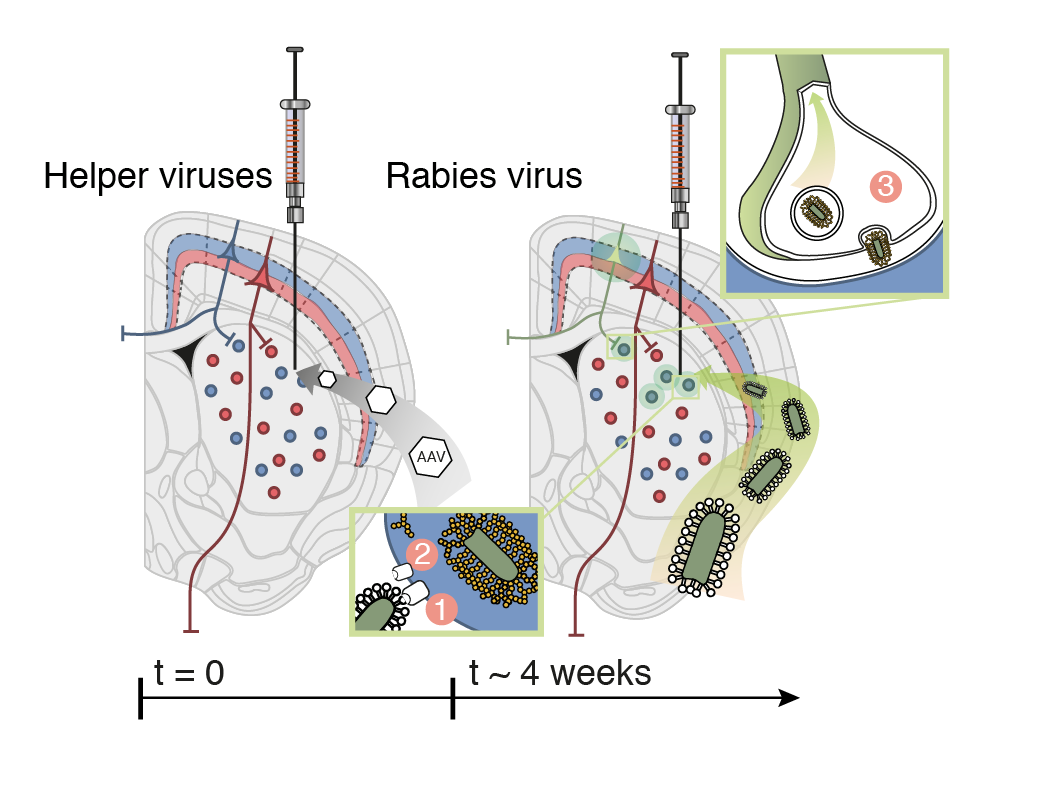
Barcoding connections
What is needed?
A method that:
- uniquely labels single postsynaptic neurons
- uniquely labels their presynaptic neurons
- uniquely label axons and trace them
low MOI barcoded rabies virus
low MOI barcoded rabies virus
impossible with todays technology
Barcoding connections

mesoscale
connectivity
31'295 neurons
single neuronal projections
+target
+strength
MAPseq: barcoded sindbis virus
for projection mapping
Barcoding connections
Barcoding connections
Sindbis virus (Togaviridae)
(+)ssRNA 9.7-11.8 kb


Cytoplasmic replication
-
Non-structural polyprotein
- processed by host and viral proteases
-
Structural polyprotein
- expressed through a subgenomic mRNA
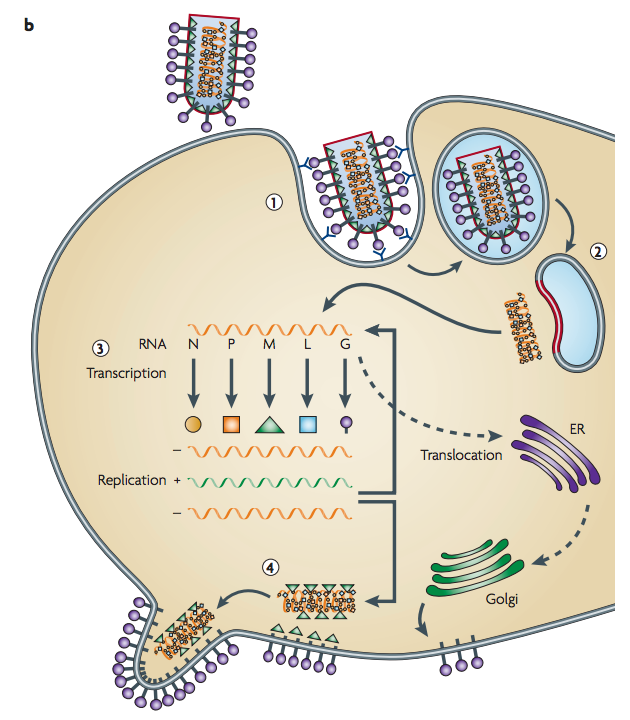
Barcoding connections
SAD B19 (Rhabdoviridae)
(-)ssRNA 11-15 kb
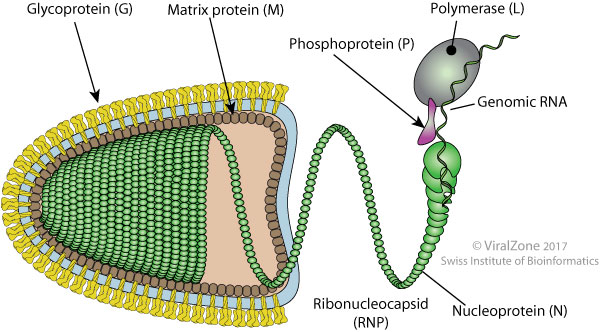

Cytoplasmic replication
transcription gradient
Barcoding connections
SAD B19 (Rhabdoviridae)
(-)ssRNA 11-15 kb

Cytoplasmic replication
located to Negri Bodies
G RABV
N RABV
Ménager et al. 2009; Lafon et al. 1985
Barcoding connections