Interrogating structure and function
using whole-brain reconstruction
Daniel Fürth
Meletis Lab
Half-time seminar
7th March 2016
daniel.furth@ki.se
understanding behavior
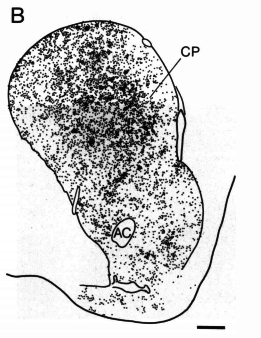
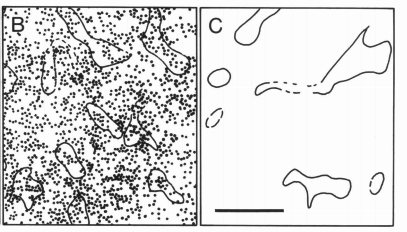
striatal models
Graybiel, A. et al. (1990) PNAS
cfos
cocaine
saline
striatal models
Graybiel, A. et al. (1990) PNAS
cfos
striatal models
Albin, Young & Penney, 1989
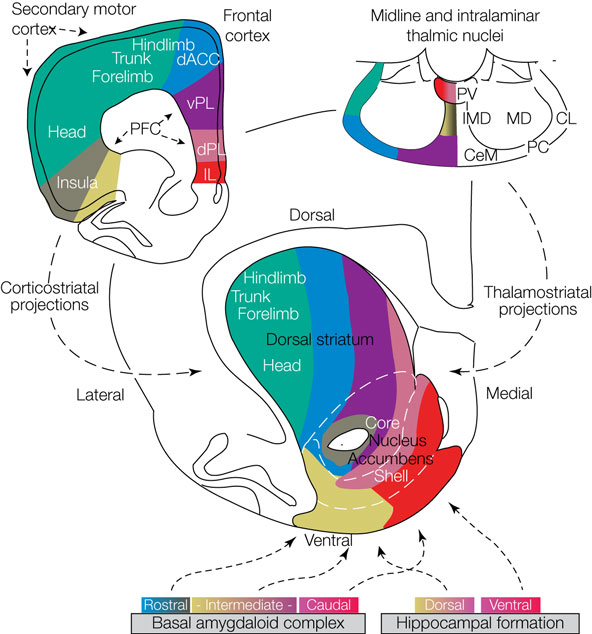

Alexander, DeLong & Stric 1986
computation in the striatum
McCulloch & Pitts, 1943
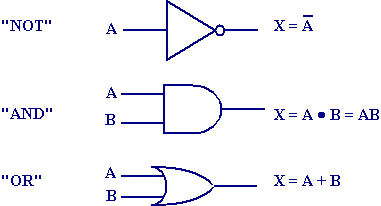

computation in the striatum
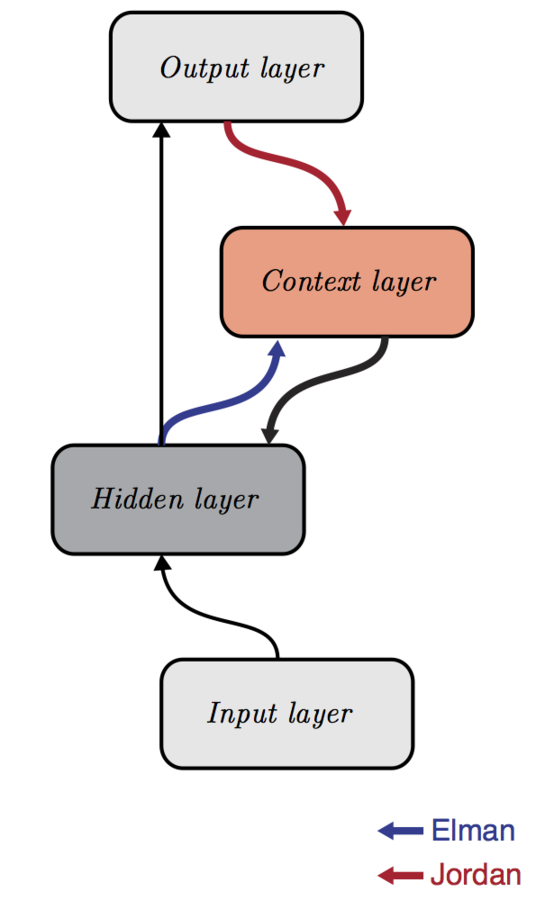
McClelland & Rumelhart 1986
Elman 1990
Jordan 1997
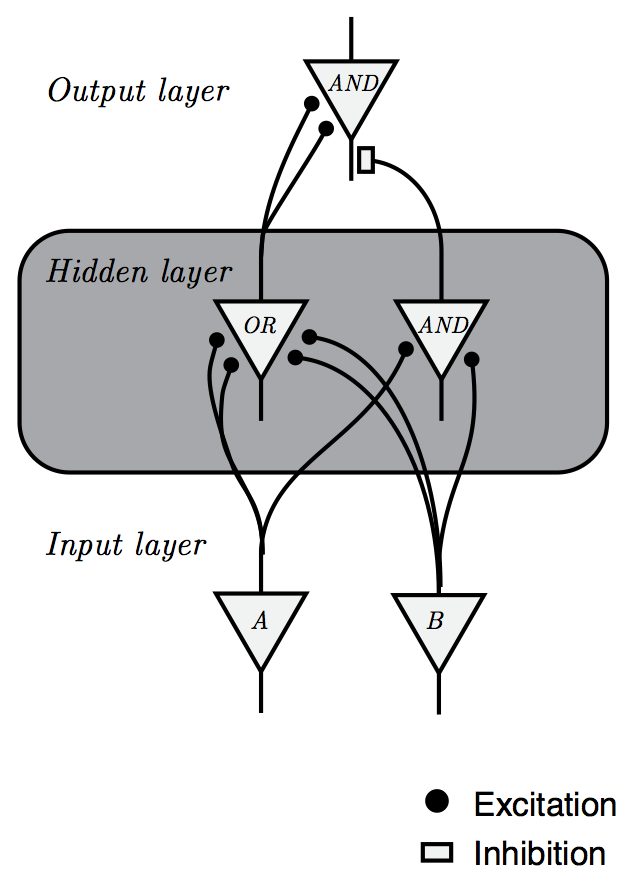
Minsky & Papert, (1972)
striatal models
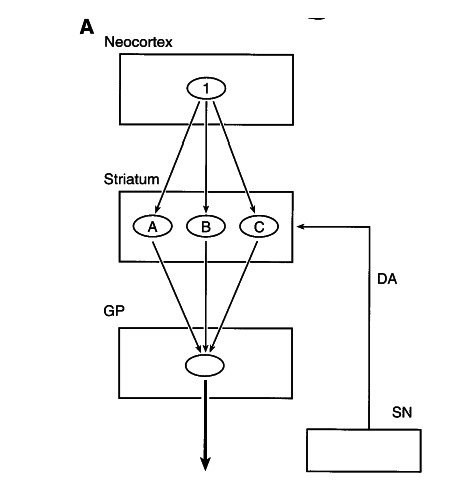
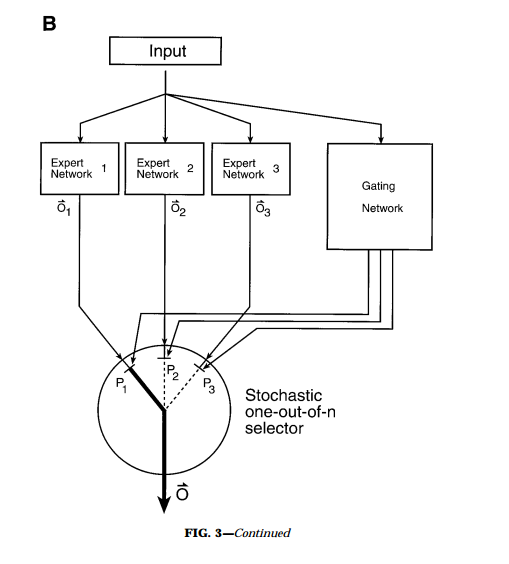
Graybiel, A. et al. (1990)
Jordan, M. I. et. al. (1991). Adaptive mixtures of local experts.
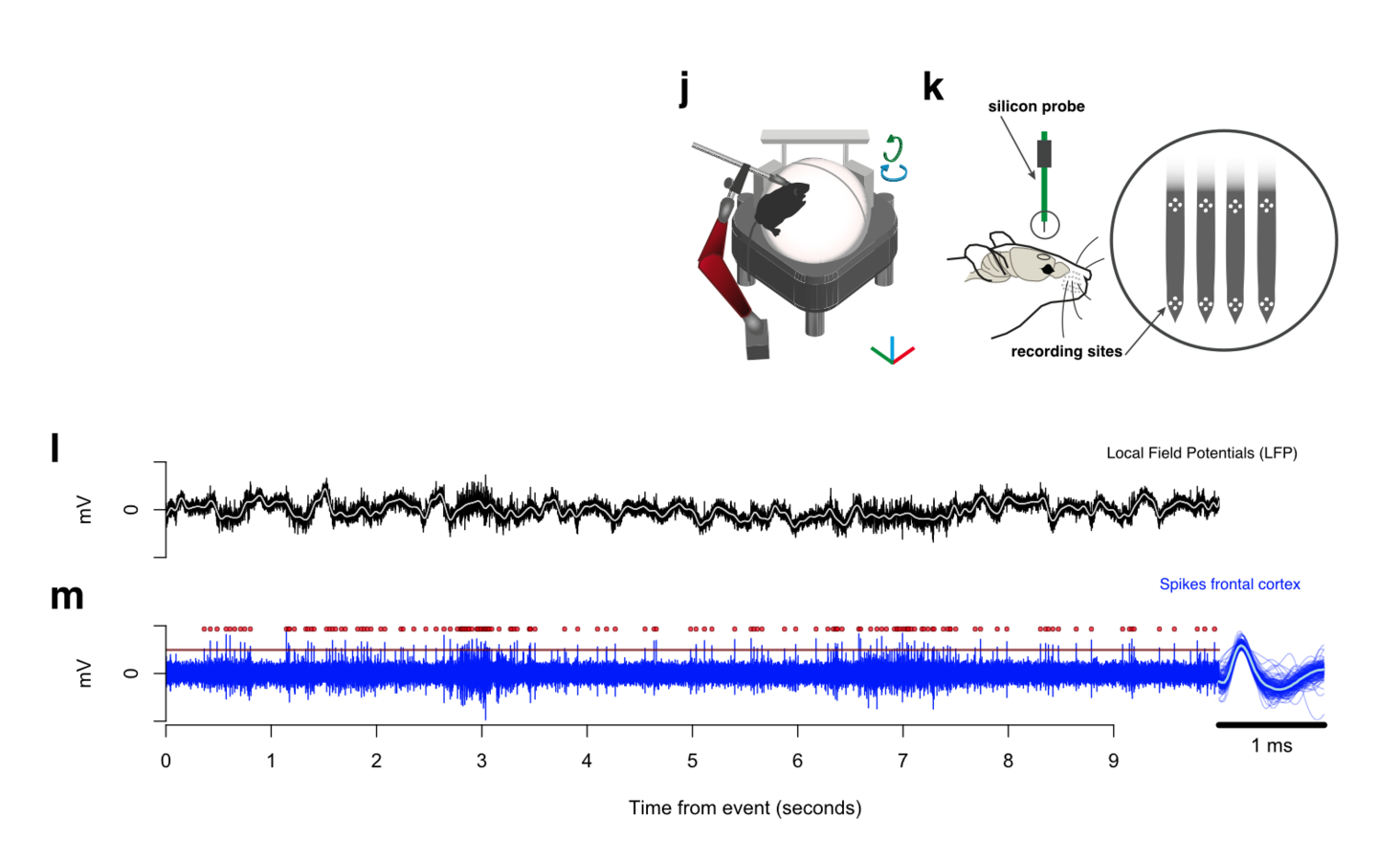
Tracing the network
The Measurement Problem
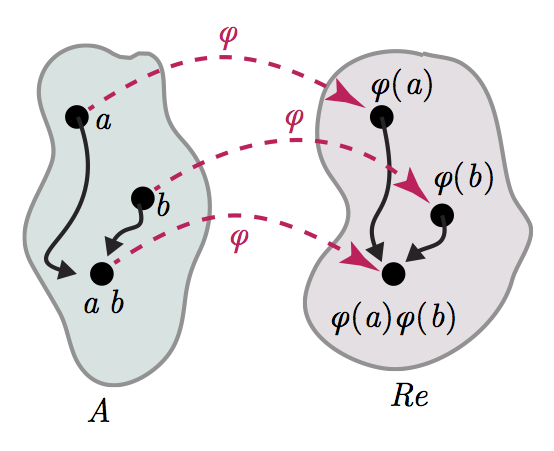
- Representational theorem
- Uniqueness theorem
The Measurement Problem
Winter & McClelland (1978)

The Measurement Problem
Winter & McClelland (1978)

The Measurement Problem

Levels of Measurement
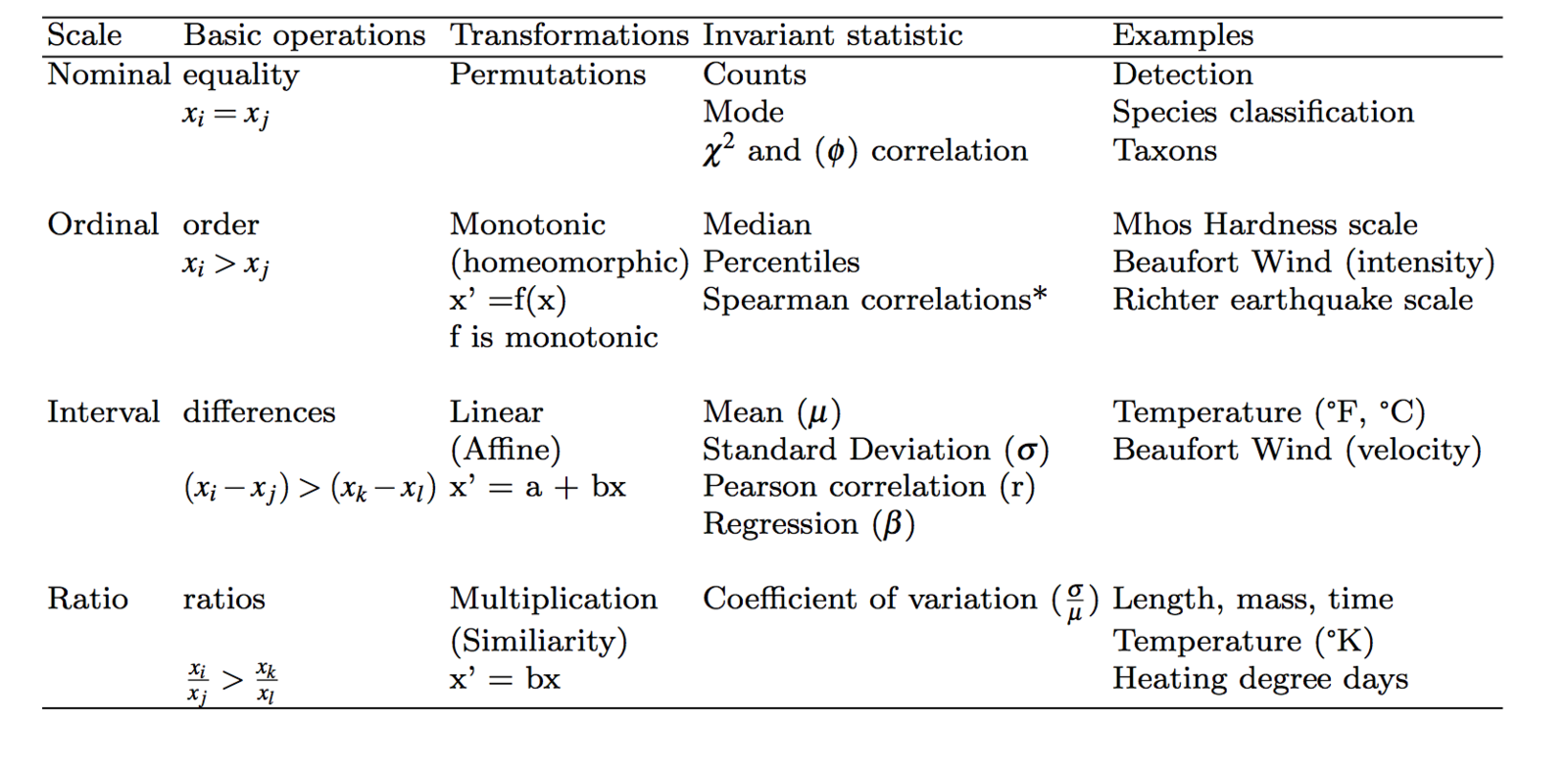
Stevens (1946)
Labeling and Counting
Church (2015)
-
Cell types
-
Connections
-
Connections strengths and types
-
Developmental lineages
-
Histories of electrical activity patterns over time
-
Histories of molecular changes over time

Tracing the network
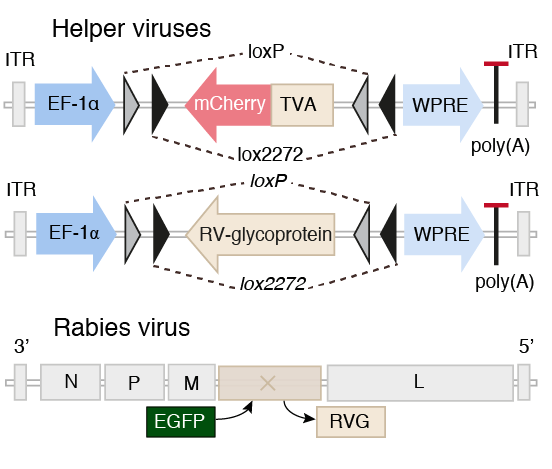
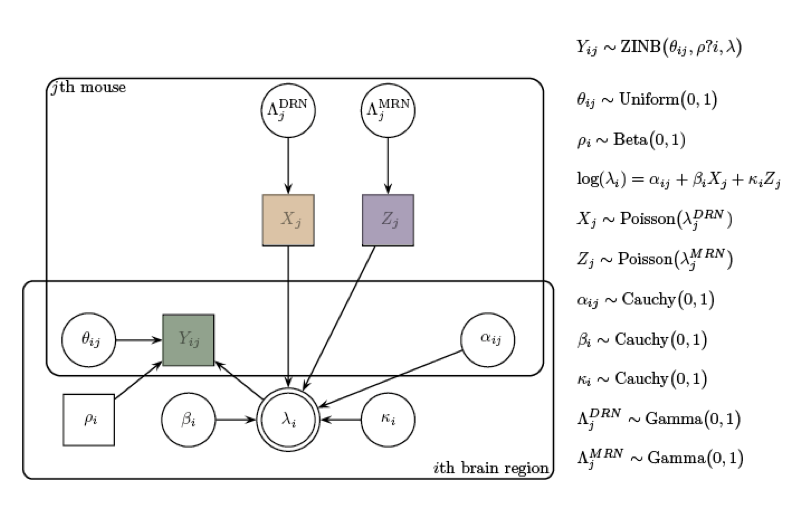
Pollak et al. 2014
www.mcstan.org

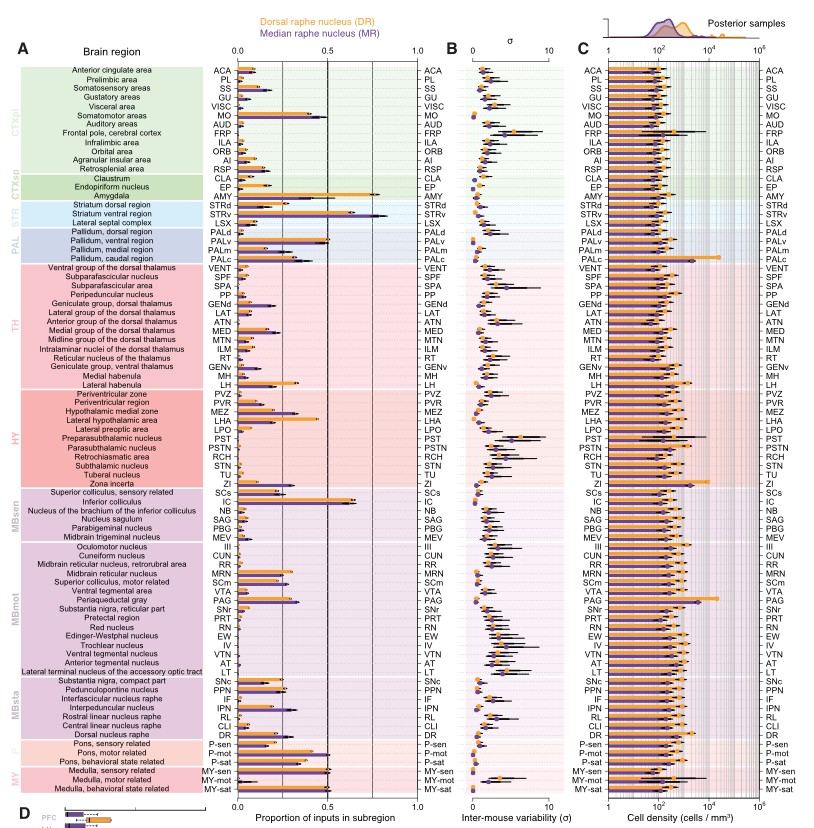
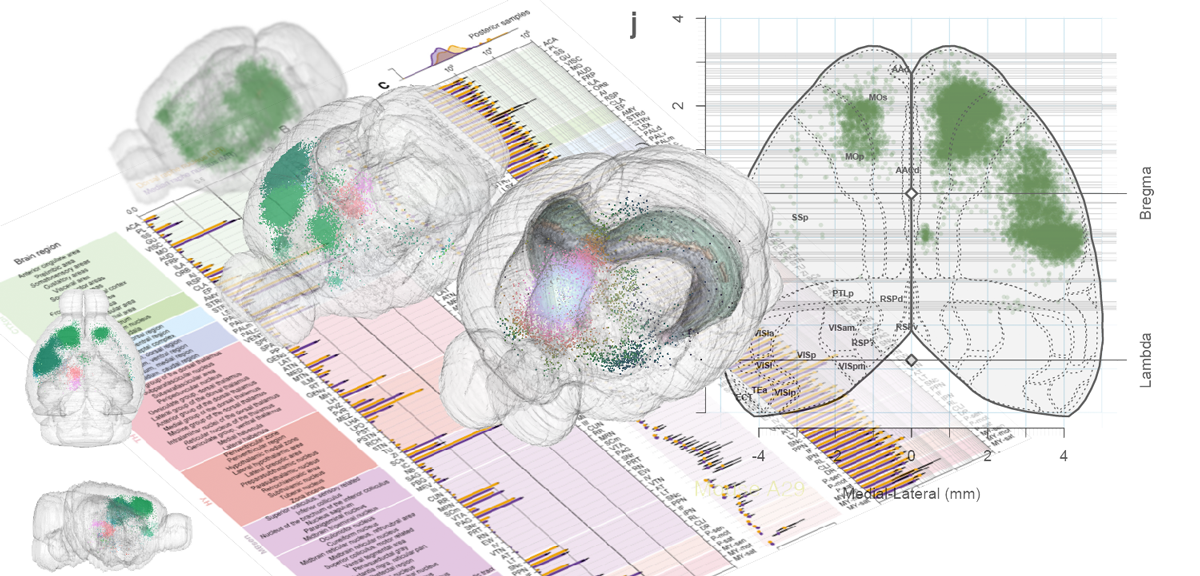
Whole-Brain Reconstruction
Pollak Dorocic et al. 2014



Reconstructing brain from sectioned tissue
Tracing the network
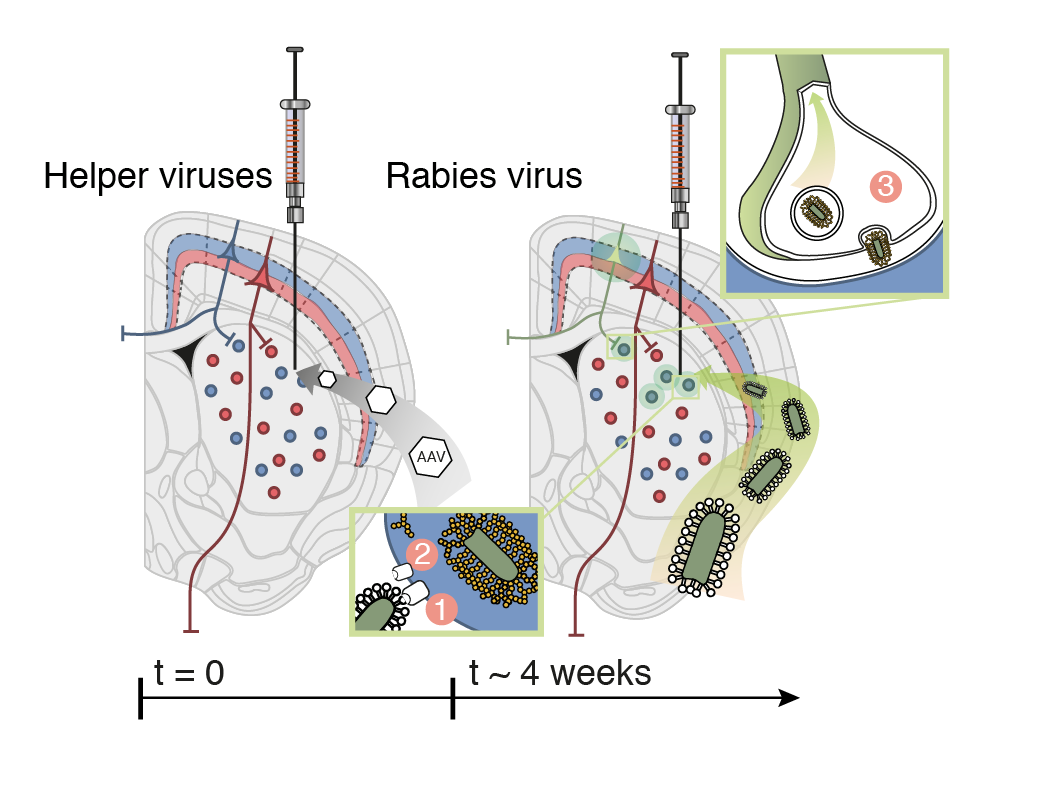
DRD2 film
56,710 neurons
Tracing the network

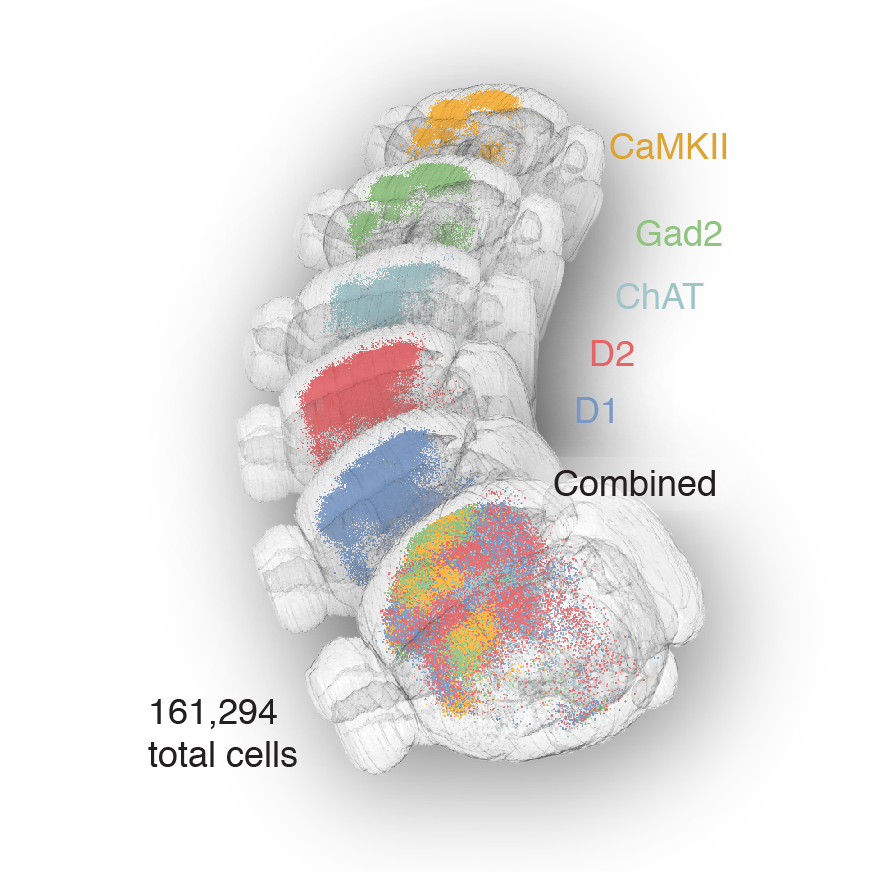
161,294 neurons film time
Tracing the network
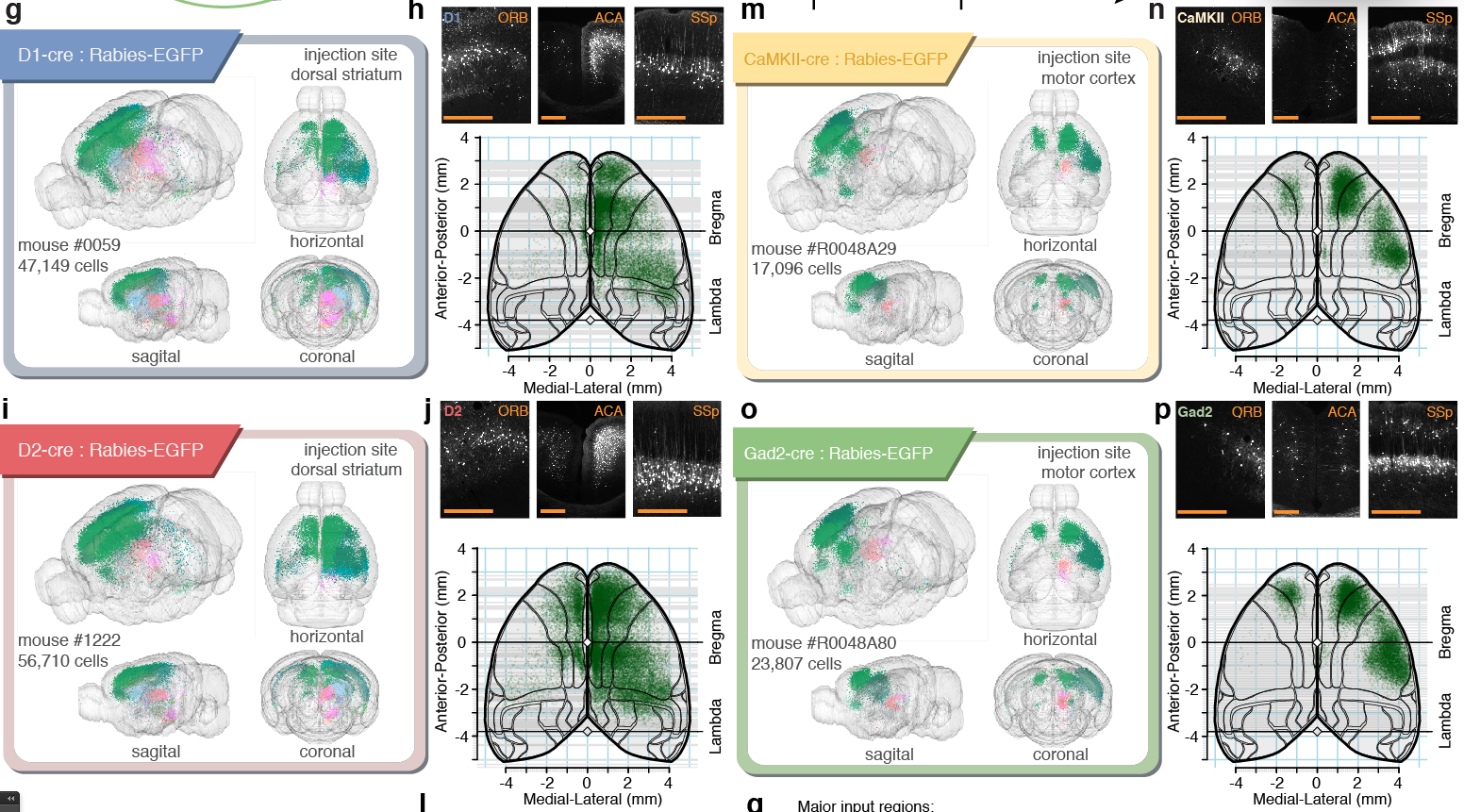
Tracing the network
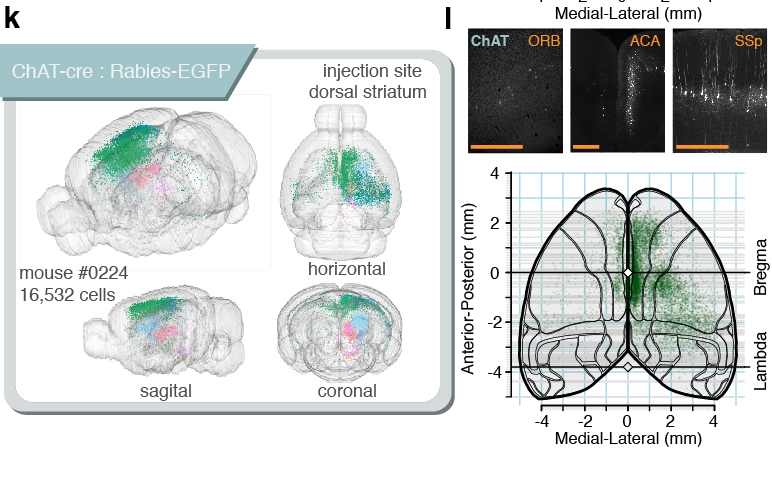
Tracing the network
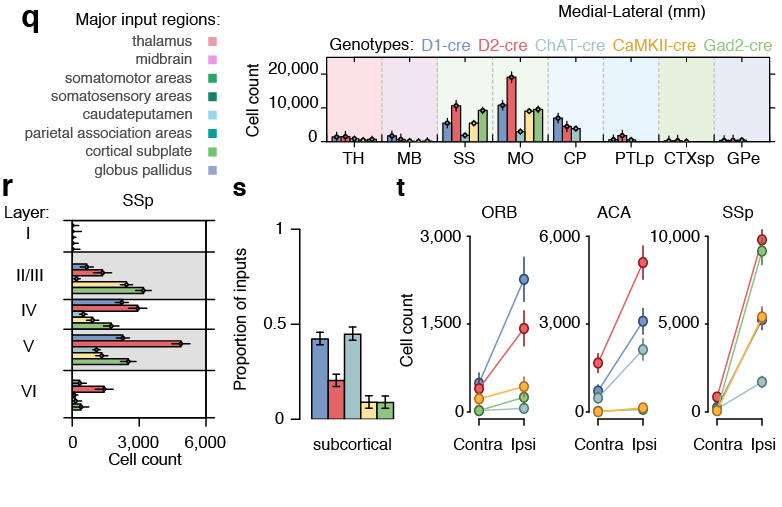
'Google maps' of neuroanatomy
'Google maps' of neuroanatomy
'Google maps' of neuroanatomy



similar to...
works with...
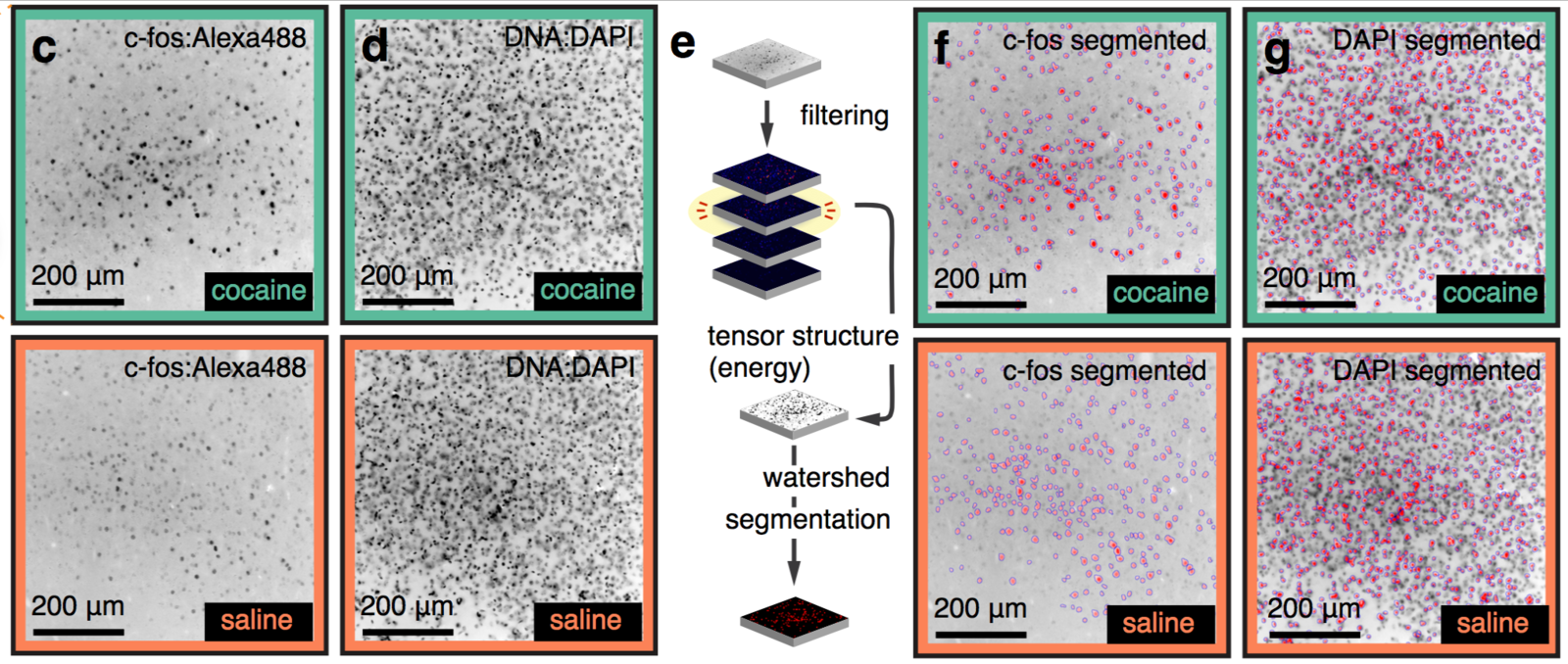
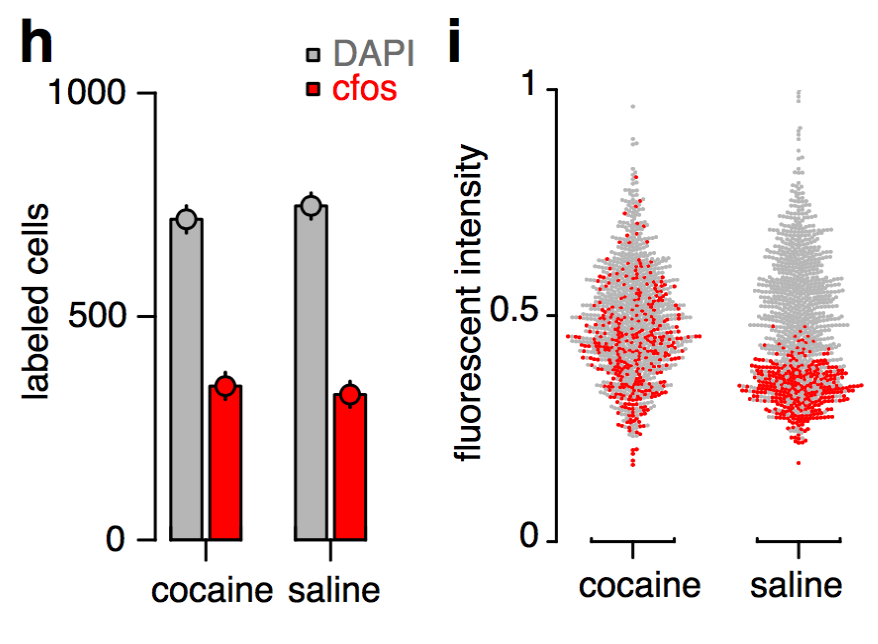
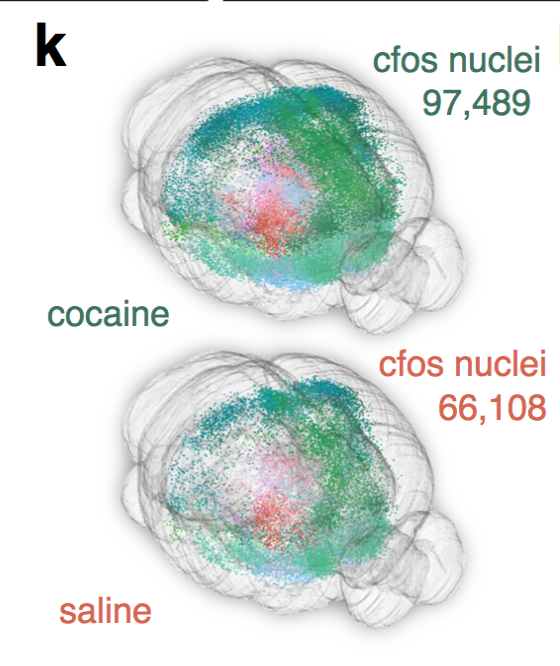
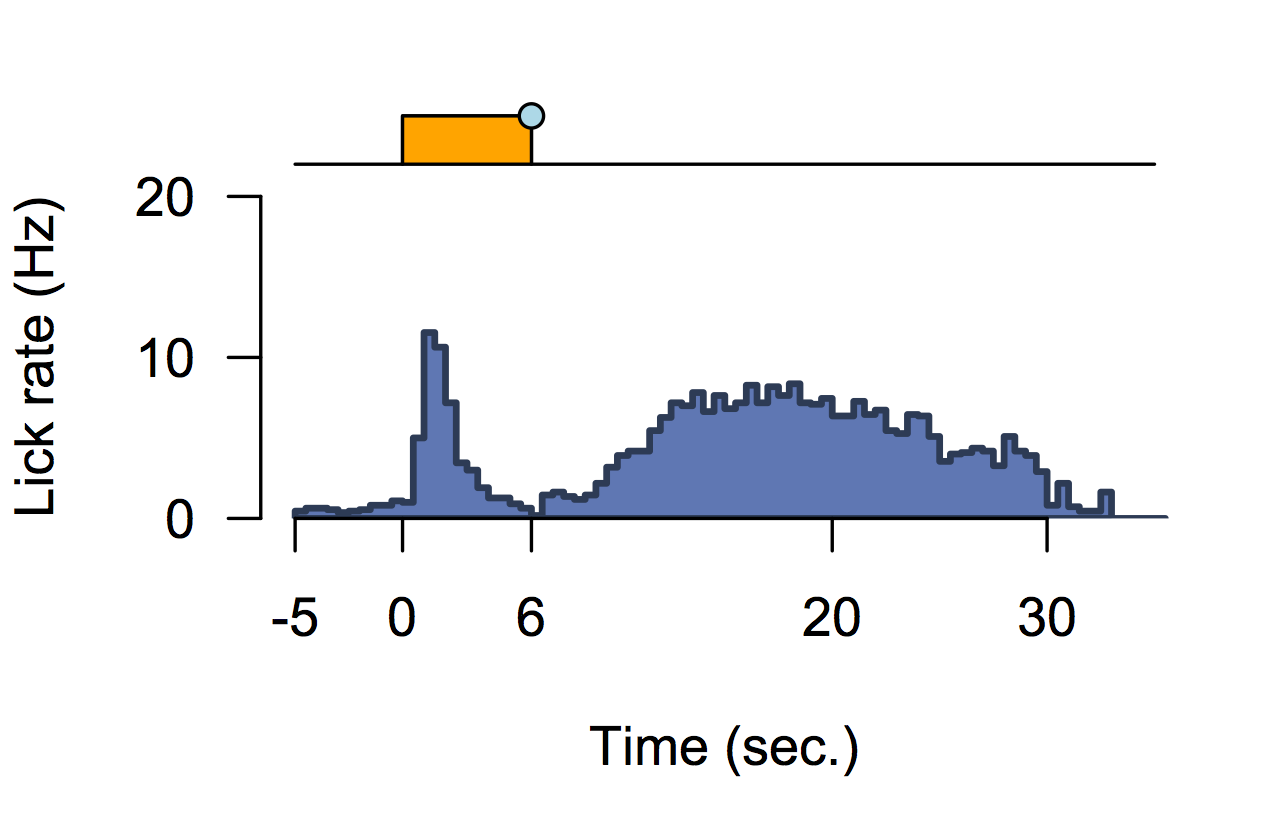
Cocaine induced locomotoric activity
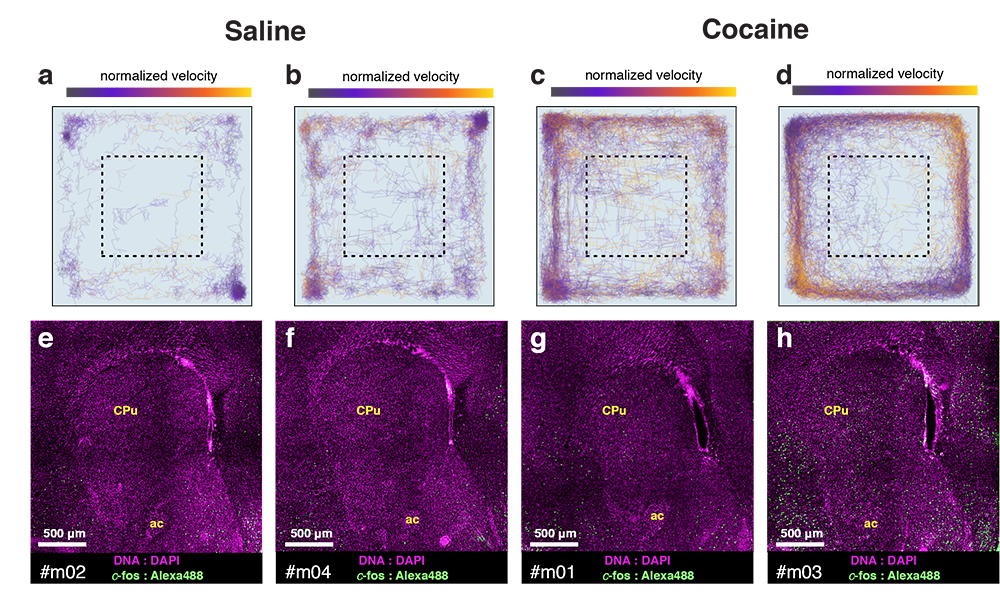
Whole-brain behavioral c-Fos mapping
Basic idea:
A
B
C
Independent
variable
Dependent
variable
Mediator
variable
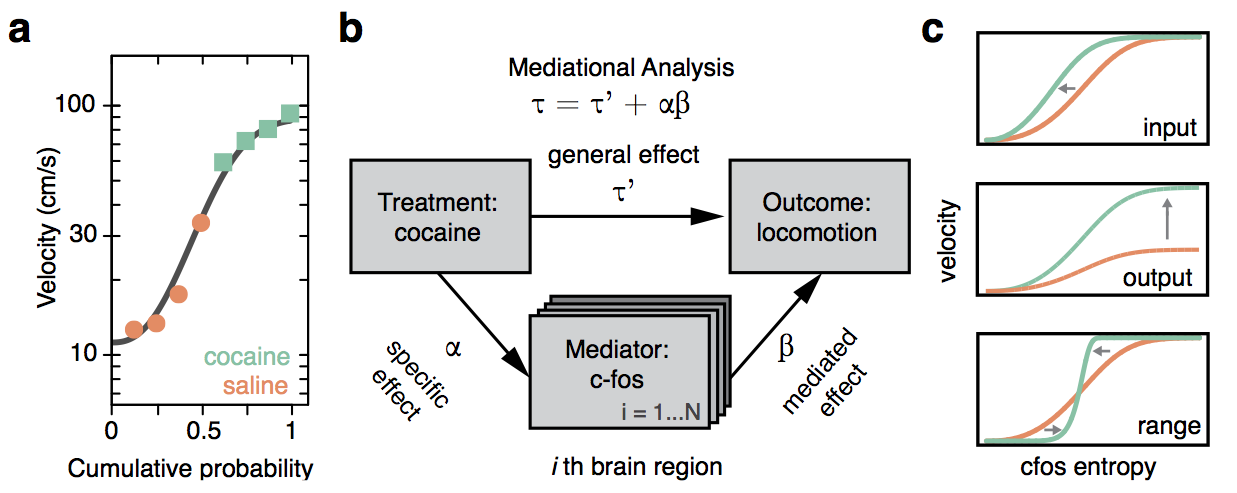
Mediational statistical analysis
Whole-brain behavioral c-Fos mapping
Basic idea.
A
B
C
cocaine
dosage
(mg/ml)
Behavior
total track length (cm)
c-fos expression
(ith region)
direct effect
indirect effect
mediating effect
Question: How much of the behavioral variability is explained by variability in c-fos expression?
-
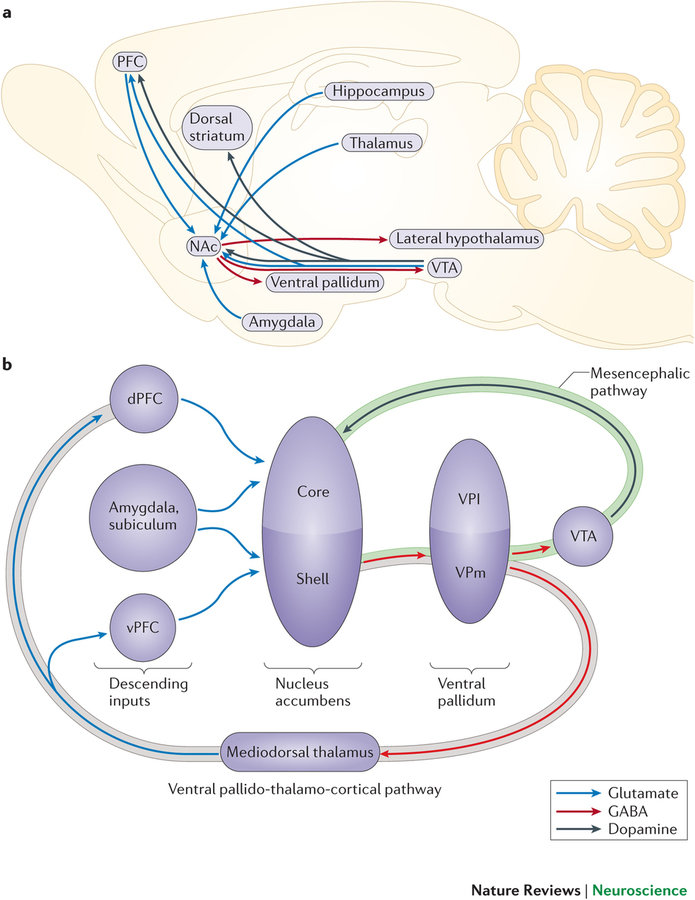
Regions of particular Interest
- NAc
- PFC
- Dorsal striatum
- Ventral pallidum
- Amygdala
- VTA
- LH
Kourrich, Calu & Bonci (2015)
Whole-brain behavioral c-Fos mapping
Whole-brain behavioral c-Fos mapping
Multiresolution decomposition
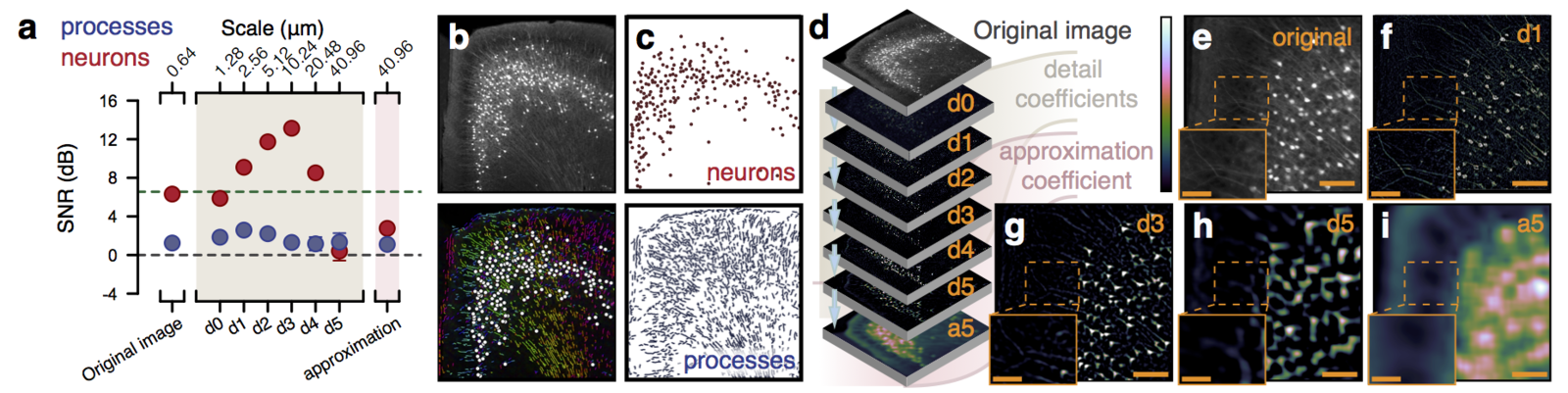
Multiresolution decomposition
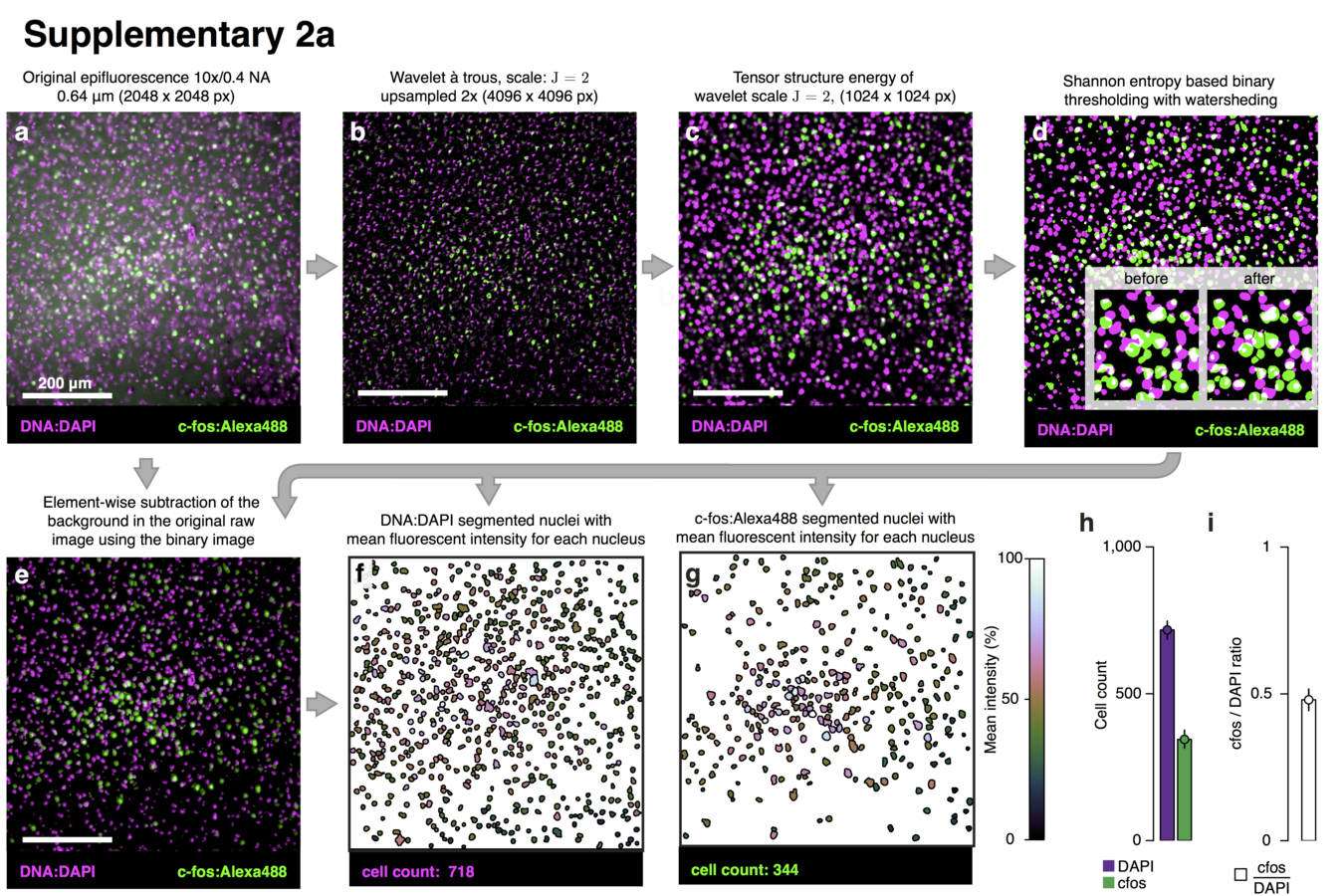
Multiresolution decomposition
Multiresolution decomposition
Functional
Can be used to segment processes and their direction.
scRNA-seq
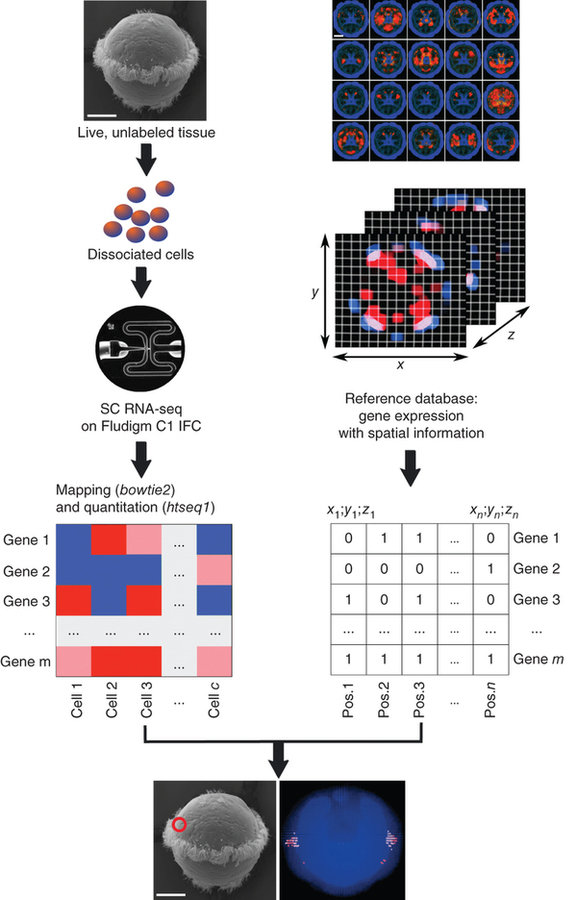
NATURE BIOTECHNOLOGY | COMPUTATIONAL BIOLOGY | ANALYSIS
High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin
Kaia Achim, Jean-Baptiste Pettit, Luis R Saraiva, Daria Gavriouchkina, Tomas Larsson, Detlev Arendt & John C Marioni
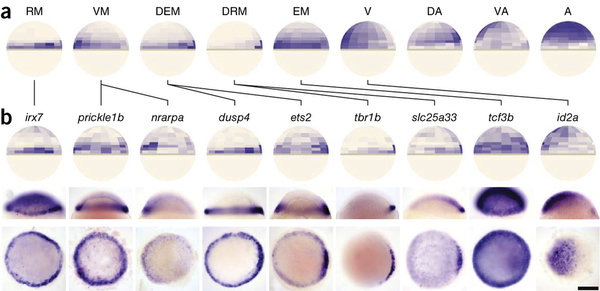
scRNA-seq
Allen Brain Reference Atlas

-
Atlas 2007 (manually drawn Nissl):
- 200 μm thick coronal sections.
-
Atlas 2011:
- 100 μm both coronal and sagital
- Atlas 2014 (connectivity avrg template)
-
Atlas 2015 (beginning of june):
- 10 x 50 μm
-
Registration atlas:
- 25 x 25 μm
-
Grid expression ISH:
- 200 x 200 μm MetaIOimage (.raw, .mhd)
scRNA-seq
Allen Brain Reference Atlas
scRNA-seq
scRNA-seq
Anatomic Gene Expression Atlas
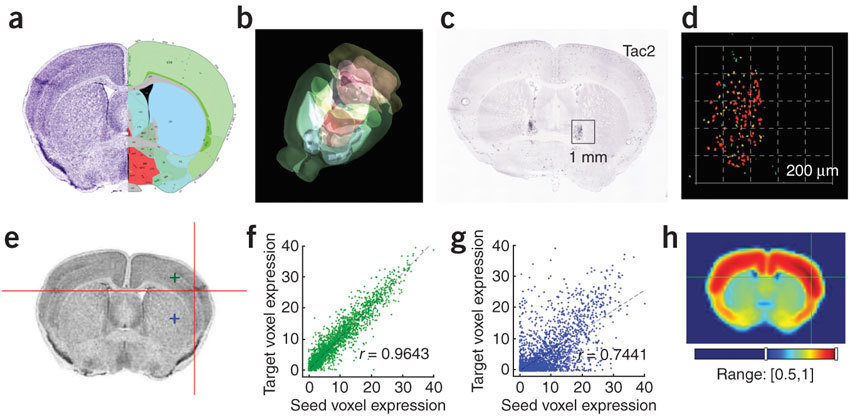
Lydia Ng, et al. (2009) Nat. Neuro.
http://mouse.brain-map.org/agea
scRNA-seq
324 cells from cortico-striatal section
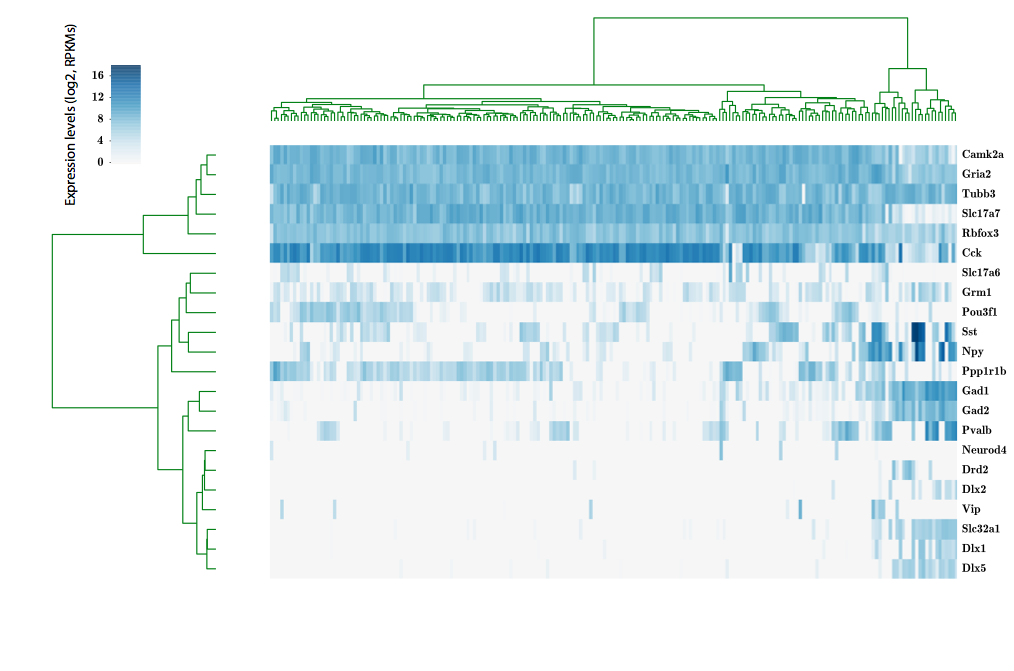
scRNA-seq
Our approach
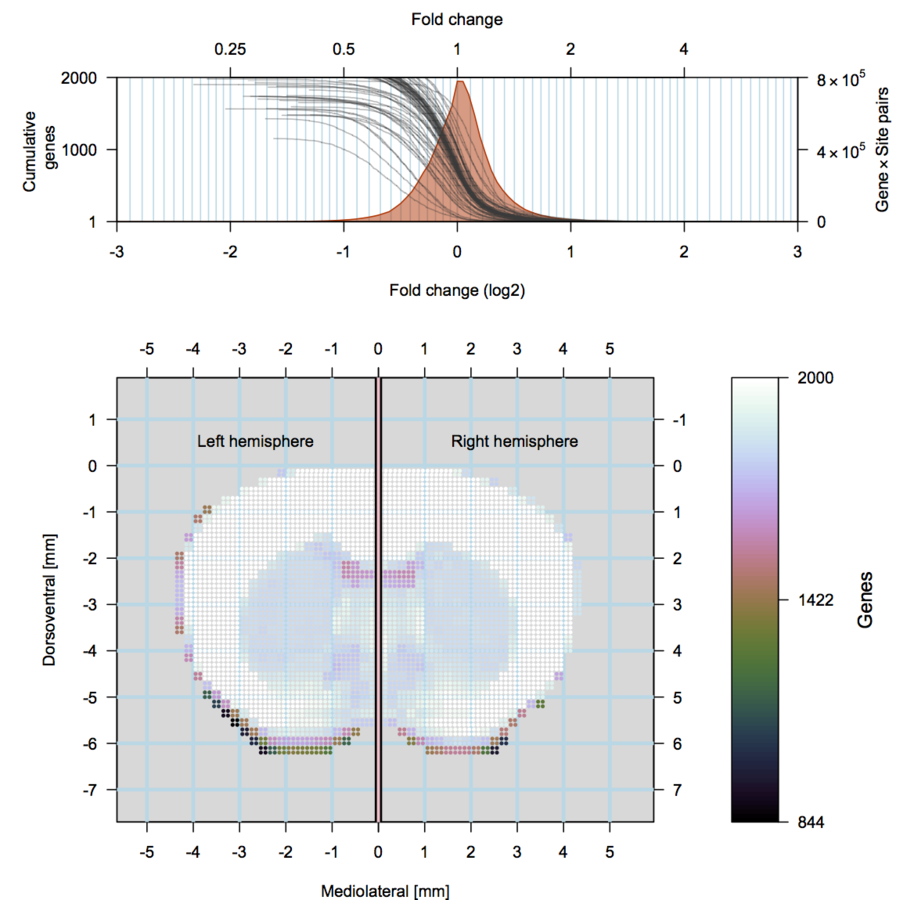
scRNA-seq
Our approach

scRNA-seq
Allen Brain Reference Atlas

-
Atlas 2007 (manually drawn Nissl):
- 200 μm thick coronal sections.
-
Atlas 2011:
- 100 μm both coronal and sagital
- Atlas 2014 (connectivity avrg template)
-
Atlas 2015 (beginning of june):
- 10 x 50 μm
-
Registration atlas:
- 25 x 25 μm
-
Grid expression ISH:
- 200 x 200 μm MetaIOimage (.raw, .mhd)
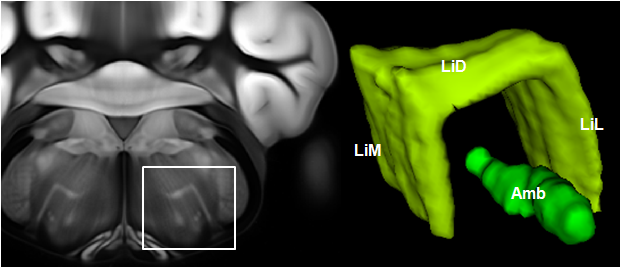
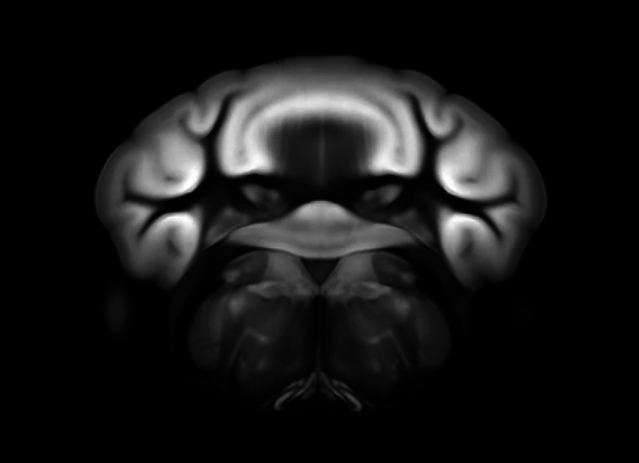
Connectivity average template (Ng et al. 2014)
scRNA-seq
Allen Brain Reference Atlas

-
Atlas 2007 (manually drawn Nissl):
- 200 μm thick coronal sections.
-
Atlas 2011:
- 100 μm both coronal and sagital
- Atlas 2014 (connectivity avrg template)
-
Atlas 2015 (beginning of june):
- 10 x 50 μm
-
Registration atlas:
- 25 x 25 μm
-
Grid expression ISH:
- 200 x 200 μm MetaIOimage (.raw, .mhd)



Connectivity average template (Ng et al. 2014)
CLARITY
Do we really have a 'BigData' problem in neuroscience?

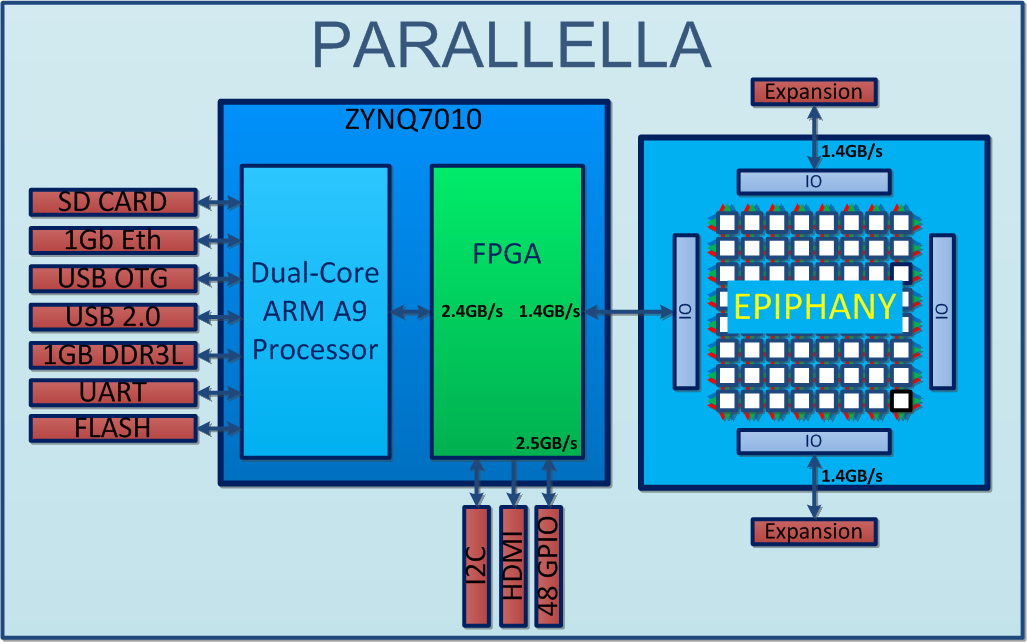
http://www.parallac.org/
10 computers (146 processors)
Up to 64 cores per processor!

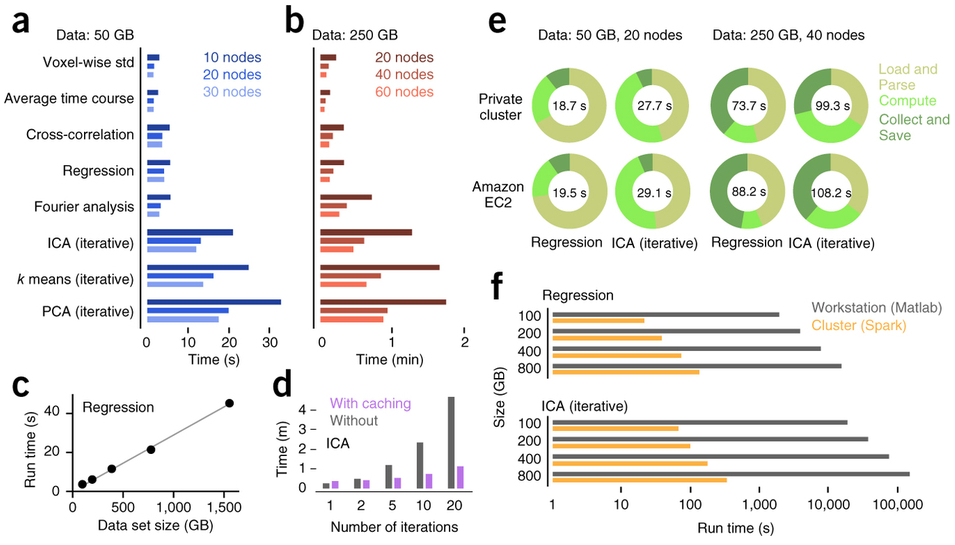
Freeman et al. (2014) Nature Methods
R package
- Why R?
- Standard data analysis:
- load some data
- estimate the density distribution.
- plot it
- Standard data analysis:
xx <- faithful$eruptions
fit <- density(xx)
plot(fit)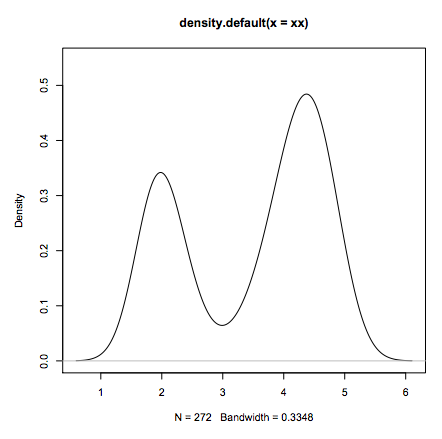
R package
- Why R?
#Line 1: loading
xx <- faithful$eruptions
#Line 2: estimate density
fit1 <- density(xx)
#Line 2: draw 10'000 bootstraps
fit2 <- replicate(10000, {
x <- sample(xx,replace=TRUE);
density(x, from=min(fit1$x), to=max(fit1$x))$y
})
#Line 3: compute 95% error "bars"
fit3 <- apply(fit2, 1, quantile,c(0.025,0.975))
#Line 4: plot the estimate
plot(fit1, ylim=range(fit3))
#Line 5: add estimation error as shaded region
polygon(c(fit1$x,rev(fit1$x)), c(fit3[1,], rev(fit3[2,])), col=’grey’, border=F)
#Line 6: add the line again since the polygon overshadows it.
lines(fit1)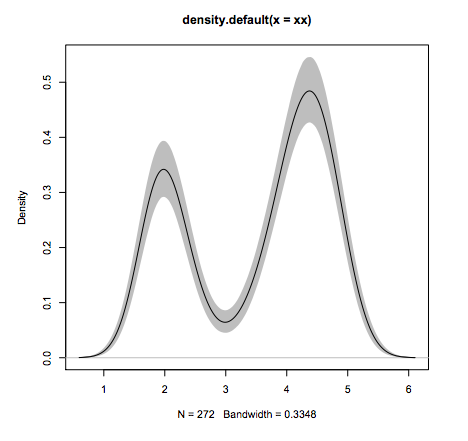
What other language can do this in 6 lines of code?
Parallel computing
- Parallel computing is extremely simple to implement from R.
# install.packages('foreach'); install.packages('doSNOW')
library(foreach)
library(doSNOW)
cl <- makeCluster(2, type = "SOCK")
registerDoSNOW(cl)
getDoParName()
#matrix operators
x <- foreach(i=1:8, .combine='rbind', .packages='wholebrain' ) %:%
foreach(j=1:2, .combine='c', .packages='wholebrain' ) %dopar% {
l <- runif(1, i, 100)
i + j + l
}Concurrency and parallel programming
- Multi threaded applications through .

#include <string>
#include <iostream>
#include <thread>
using namespace std;
//The functions we want to make the thread run.
void task1(string msg)
{
cout << "task1 says: " << msg;
}
void task2(string msg)
{
cout << "task1 says: " << msg;
}
//Main loop.
int main()
{
thread t1(task1, "Task 1 executed");
thread t2(task2, "Task 1 executed");
t1.join();
t2.join();
}Rcpp
Concurrency and parallel programming
- Multi-threaded applications through .

#include <string>
#include <iostream>
#include <thread>
using namespace std;
//The functions we want to make the thread run.
void task1(string msg)
{
cout << "task1 says: " << msg;
}
void task2(string msg)
{
cout << "task1 says: " << msg;
}
//Main loop.
int main()
{
thread t1(task1, "Task 1 executed");
thread t2(task2, "Task 1 executed");
//let main wait for t1 and t2 to finish.
t1.join();
t2.join();
}Rcpp

Dual core
Thank you!
scRNA-seq
Gene specificity
about ~24'000 genes expressed in the brain.
