Project Cognoma
What does Project Cognoma do?
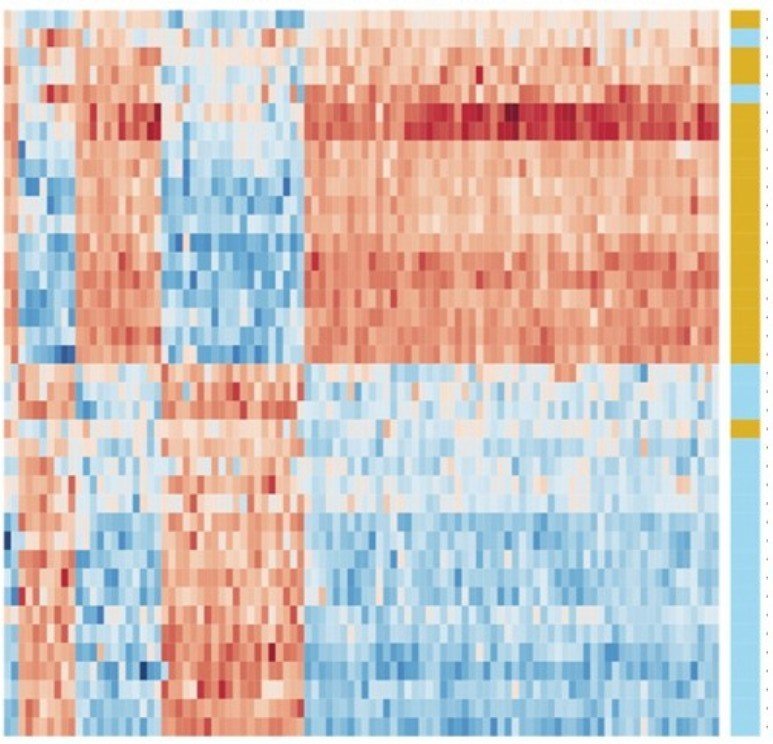
Image derived from https://doi.org/bmfx (CC BY)
genes / features / predictors / x
tumors / samples / observations
mutation / status / outcome / response /
y
Use Case from Cancer Biologist
- Robert's lab is interested in maintenance of mitochondria in cancer.
- Interested in developing a therapeutic to inhibit mitophagy. This is likely to have low toxicity.
- A priori genelist - PARK2, PINK1, NIX, BNIP3, DRP1, PARL

Goal: Find "Hidden Responders"
- Pathology samples
- Cell lines
- PDX Models
Cares about classifier performance:
Time + $$$$
Project Cognoma:
Goals
- "Putting machine learning in the hands of cancer biologists"
- Our tagline and ultimate goal
- Project Cognoma is a community project
- We all benefit from the collective expertise to build a superior product
- Open science!
- Everyone learns something new
- We will be using cutting edge tech
- Encourage cross-talk between groups and open collaboration
Current Progress on GitHub
- Our organizational & general issues repository is cognoma/cognoma on GitHub
- Introduce yourself (Issue #2)
- Project domain: cognoma.org (Issue #2)
- Biological use cases (Issues #11 & #3)
- Contribution guidelines (see GUIDELINES.md)
- Everything should go through GitHub
- Use issues liberally
- Pull request model
- Component repositories (django-cognoma and task-service)
Structure of this Datathon
- Break into groups
- Groups are assigned topics
- Feel free to redefine groups and add new topics
- Report on the GitHub at the end
- Short (~3 minute) report to the group to close the night
Tonight: Breakout Session
- Data
- Machine Learning
- Backend group
- Django
- Task Service
- Frontend group
- Javascript Webapp
- Design
- Community & Management
Data Group
- What is the ideal format of the data?
- What processing is necessary to get the data into a tidy format?
- Can we begin to explore the data?
- What does it look like?
- Can we perform some exploratory analyses?
- How should we prepare and preprocess the data? Do we need to do normalization?
- What is the licensing of the TCGA data we're using? Can we release data as CC0?
Machine Learning Group
- We need supervised machine learning algorithms for binary outcome data. There will be between 10,000 and 30,000 features (genes). There will be between 100 and 11,000 samples. The status will likely be highly unbalanced (e.g. 100 positives & 3000 negatives).
- What are possible algorithms (regression, SVM, etc)?
- How should we address overfitting?
- Should we only report cross-validated performance?
- Is scikit-learn a good package to start with?
- Can we start drafting the design of the algorithm chooser?
Backend Group
- Familiarize with prior work
- django-cognoma issues
- software architecture proposals (Issue #5)
- technical ideas (Issue #4)
- What technologies should the backend use?
- Refine the architecture
- What technologies would be good to use from a pedagogical perspective, e.g. Docker?
Frontend Group
- What technologies should the frontend use?
- How should we design the javascript webapp
- What are the best ways to coordinate frontend development?
- How can we make a GUI query builder for Hetionet so researchers can identify a set of genes even if they don't know Cypher?
Design Group
- Can we create a project logo? Including a favicon?
- Do we need accounts? Are there login-free methods of query preservation? See this issue.
Community & Management
- How can we ensure lot's of contribution from lot's of contributors?
- What is currently making it difficult to contribute or join the project?
- How is the best way to coordinate and manage the community?
- How do we make sure everyone learns something?
Happy Hacking!
See these slides at:
https://slides.com/dhimmel/cognomathon
