Hetnets + Biomedicine + Neo4j = Hetionet
Slides at slides.com/dhimmel/datathon
Daniel Himmelstein (@dhimmel)
Greene Lab at Penn (@GreeneLab)
Can we make a machine learn all biomedical knowledge?
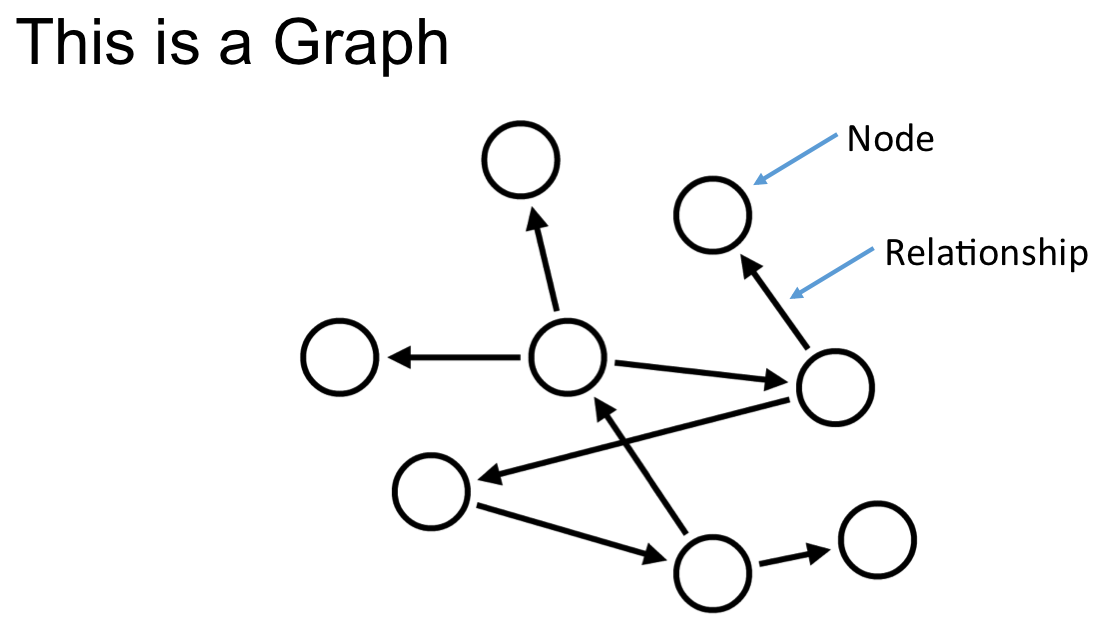
Networks encode knowledge…
Hetnets encode diverse knowledge
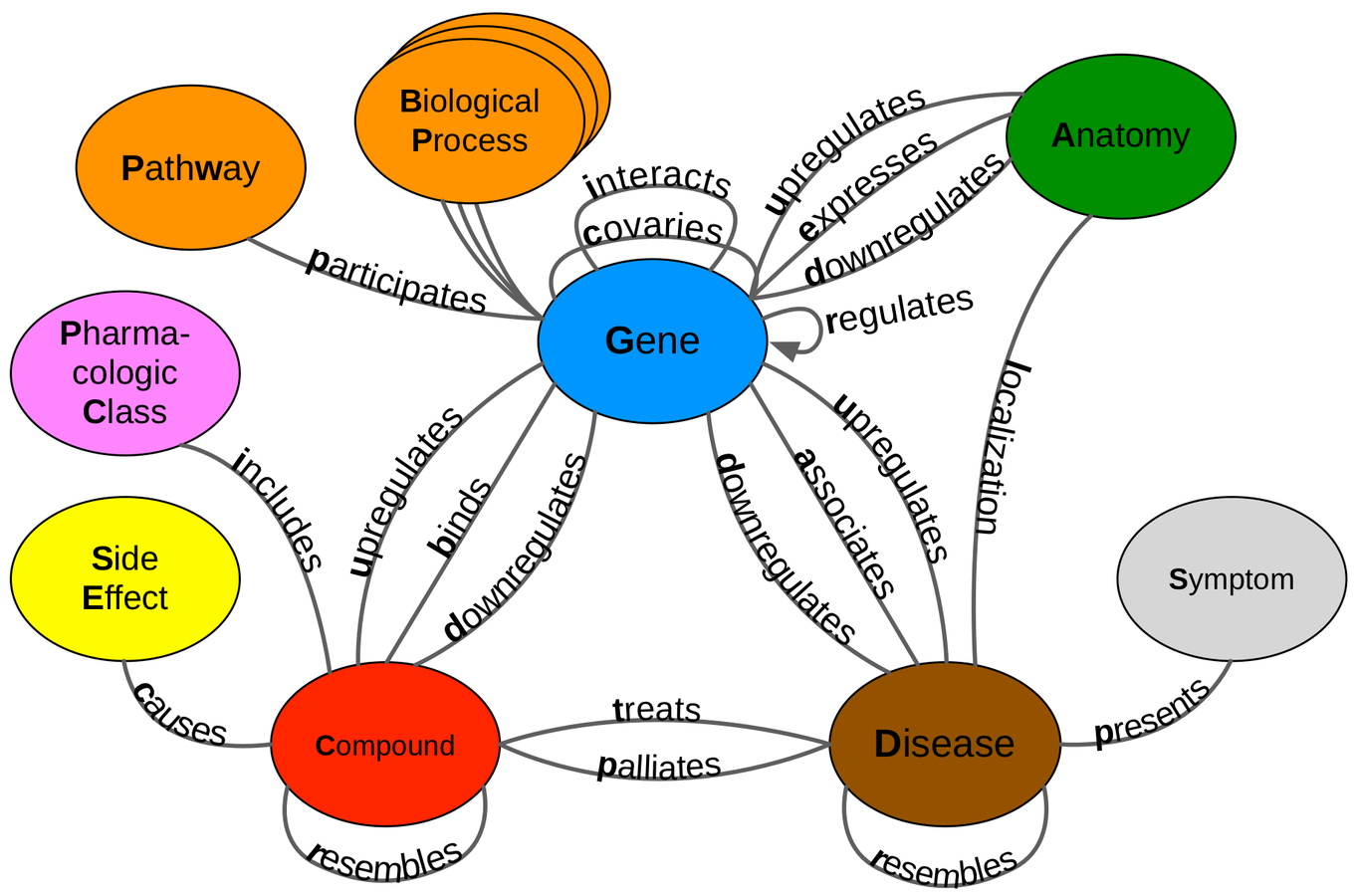
Gene
User
Page
Junction
Compounds
Diseases
Pharmacologic Classes
Side Effects
Symptoms
Anatomies
Genes
Pathways
Molecular Functions
Biological Processes
Cellular Components
-
1,552 small molecule compounds
-
137 complex diseases
-
755 treatments
-
47,031 nodes of 11 types
- 2,250,197 edges of 24 types
- 29 resources
- millions of studies from last half century
- Hetionet integrates data from 29 resources
- 12 had an open license
- 9 had no license
-
Incompatibilities - Share Alike vs Non-Commercial
- Requested permission for 11 resources
- Median time to first reponse was 16 days
-
2 affirmative responses
- Removed MSigDB
- "LICENSEE agrees not to put … the DATABASE on a … server … that may be accessed by any individual other than the LICENSEE."
- LICENSEE agrees to provide … a written evaluation of the PROGRAM and the DATABASE, including a description of its functionality or problems and areas for further improvement
Legal barriers to data reuse
Recommendation:
release data under an open license
Slide courtesy of Nicole White (@_nicolemargaret)
Slide courtesy of Nicole White (@_nicolemargaret)
Slide courtesy of Nicole White (@_nicolemargaret)
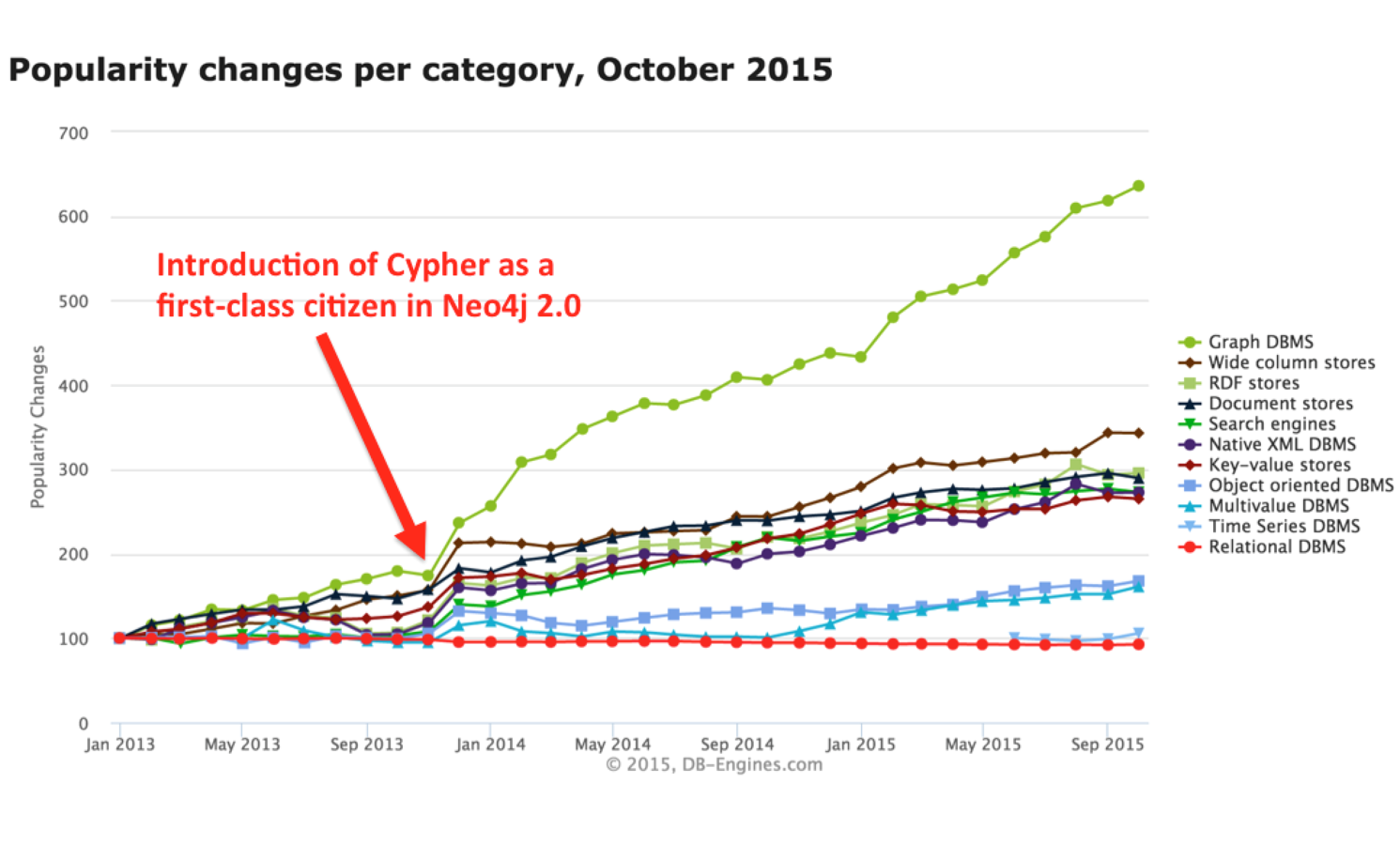
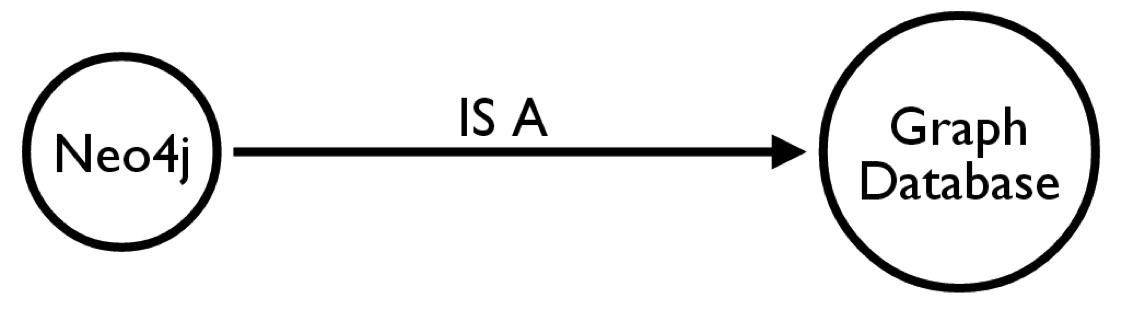
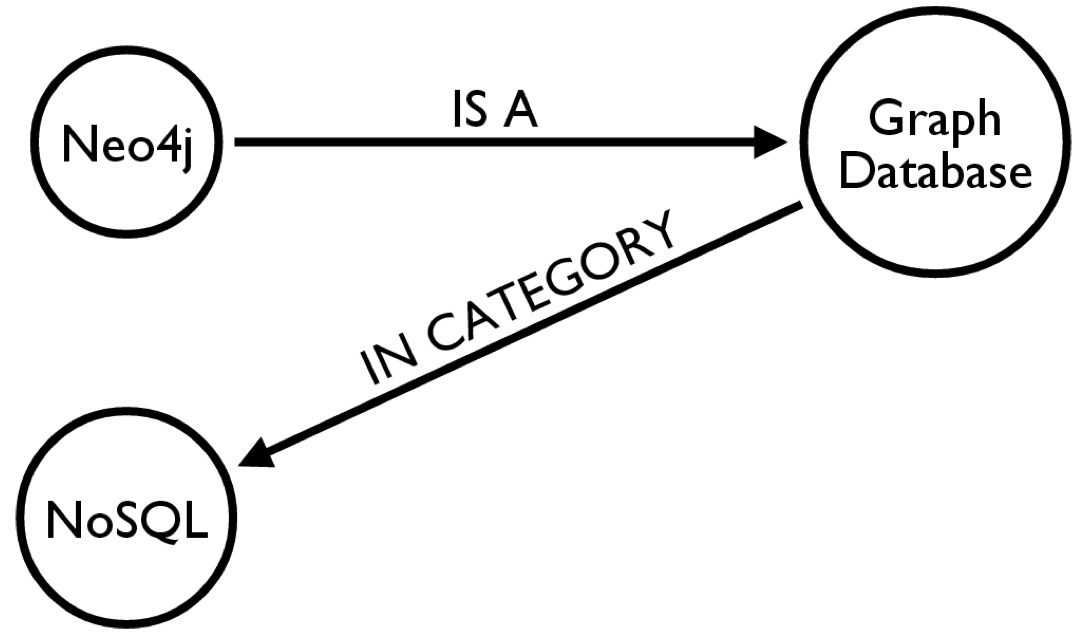
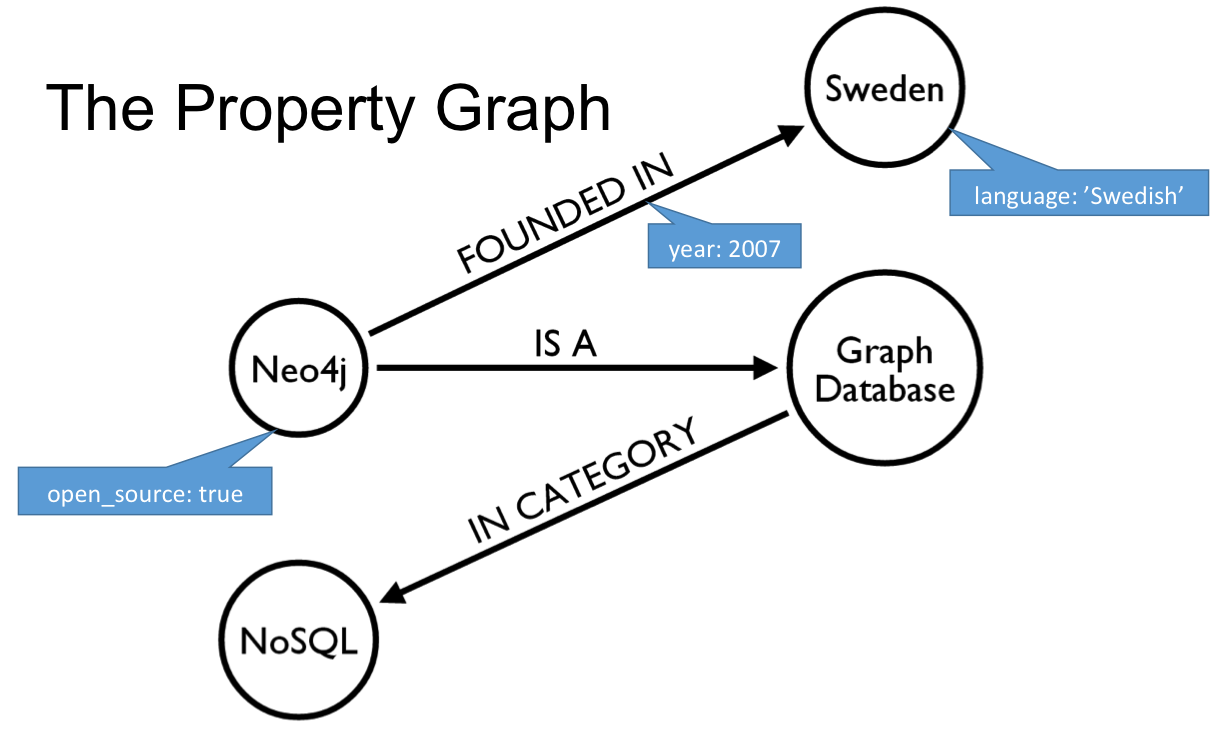
Cypher Query Language
So what does Hetionet know…
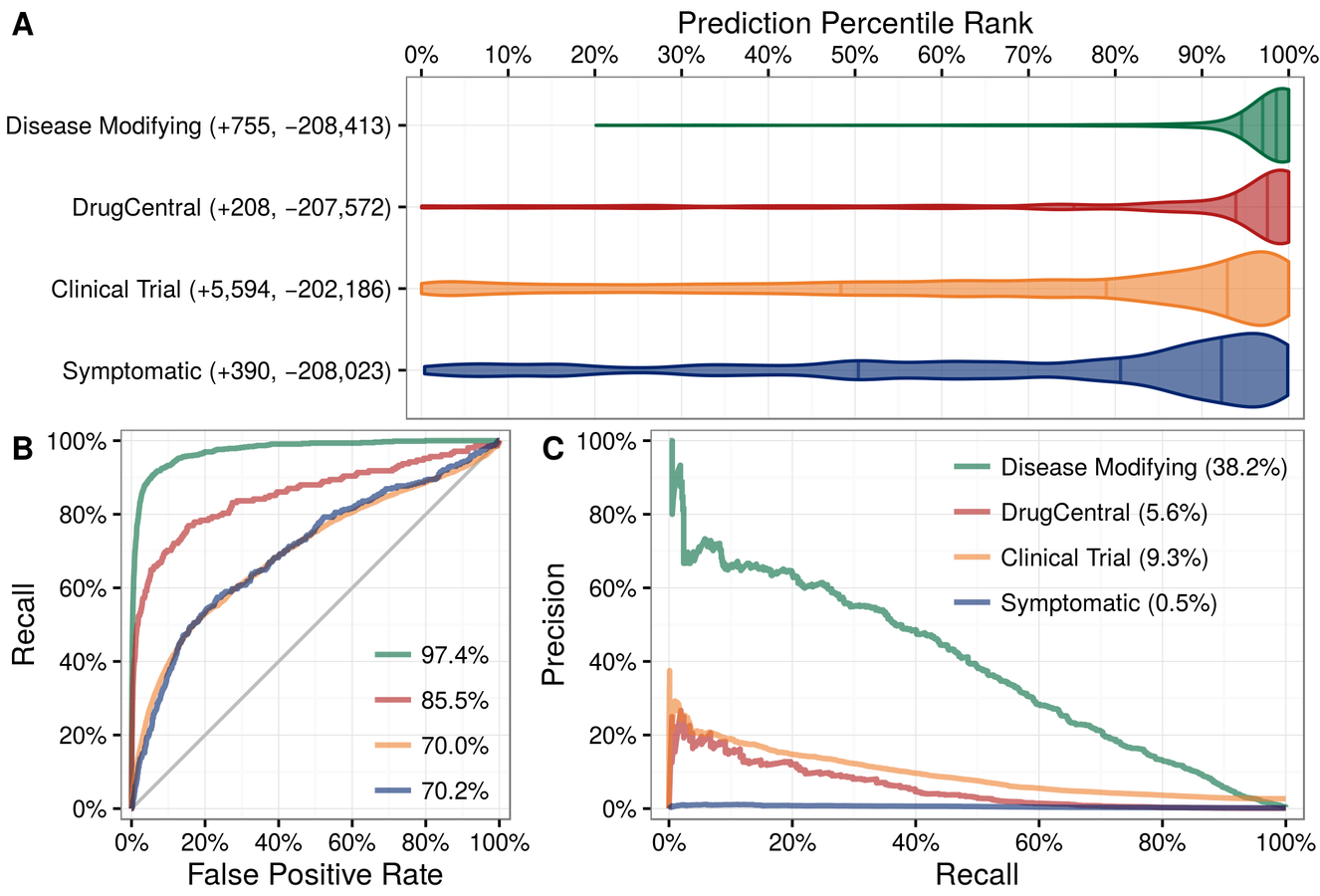
Can we predict new uses for existing drugs?
209,168 Predictions (het.io/repurpose)
Hetionet browser at neo4j.het.io
MATCH path = (n0:BiologicalProcess)
-[:PARTICIPATES_GpBP]-(n1)
-[:BINDS_CbG]-(n2:Compound)
WHERE n0.name = 'myelination'
RETURN pathCan answer versatile questions such as:
What compounds that target genes involved in myelation?
Go to neo4j.het.io & try to:
- Retrieve the Disease node named "lung cancer"
- Find which anatomies (tissue types) where lung cancer localizes (LOCALIZES_DlA)
- Find all genes associated with "spinal cancer" (ASSOCIATES_DaG)
- Find all genes associated with both "liver cancer" and "kidney cancer"
- Find all genes that participate in the "mitotic spindle checkpoint" BiologicalProcess (PARTICIPATES_GpBP)
- Find all genes that participate in the "mitotic spindle checkpoint" and are expressed in the lung (EXPRESSES_AeG)
Solution queries
MATCH (node:Disease {name: "lung cancer"}) RETURN nodeMATCH (:Disease {name: 'lung cancer'})-[rel:LOCALIZES_DlA]->() RETURN relMATCH (:Disease {name: 'spinal cancer'})-[r:ASSOCIATES_DaG]->() RETURN rMATCH path=(:Disease {name: 'liver cancer'})-[:ASSOCIATES_DaG]-(:Gene)-[:ASSOCIATES_DaG]-(:Disease {name: 'kidney cancer'}) RETURN pathMATCH ({name: 'mitotic spindle checkpoint'})-[rel:PARTICIPATES_GpBP]-() RETURN relMATCH path=(:BiologicalProcess {name: 'mitotic spindle checkpoint'})-[:PARTICIPATES_GpBP]-(gene:Gene)-[:EXPRESSES_AeG]-(:Anatomy {name: 'lung'}) RETURN path
Project Cognoma
Hetionet will be one of several components in Project Cognoma
Putting machine learning in the hands of cancer biologists
Thanks Neo4j for the pizza