Integrating biomedical knowledge to predict new uses for existing drugs
Models, Inference & Algorithms, Broad Institute
8:30 am – 9:20 am, February 22, 2017
Monadnock Room, 415 Main Street, Cambridge, MA
By Daniel Himmelstein
@dhimmel
Slides at slides.com/dhimmel/mia
Greene Lab
I'm a data scientist
http://www.greenelab.com/

Sandler Neurosciences Center

Sergio
How do you teach a computer biology? Our goal was to predict new uses for existing drugs. But we're data scientists, not pharmacologists. So we set out to encode the knowledge from millions of biomedical studies from the last half century. Using a heterogeneous network (hetnet) as our data structure, we were able to condense a large portion of biomedical knowledge into a network with 47,031 nodes of 11 types and 2,250,197 relationships of 24 types. The network is named Hetionet v1.0 and lives at https://neo4j.het.io.
Hetionet enables queries that span many types of information. While such queries were possible before Hetionet, they often took months of data integration, preprocessing, and specialized query scripts. Now complex queries can be written in minutes using the Cypher query language for hetnets. Accordingly, we were able to perform ~47 million queries to assess the connectivity between 136 diseases and 1,538 compounds. Next, we compiled a catalog of 755 disease-modifying treatments and learned which types of network paths could predict whether a compound treats a disease. In total, we predicted probabilities of treatment for 209,168 compound-disease pairs (http://het.io/repurpose). Our method also allows you to compare which types of information were valuable for predicting drug efficacy. Project Rephetio, the codename for this project, was performed openly online in realtime (https://doi.org/bszr). In total, 40 community members provided feedback across 86 project discussions.
Attend the primer to learn more about Project Rephetio & Hetionet as well as hetnets for data integration and the Neo4j graph database. Research continuous as a set of open source GitHub repositories, allowing anyone interested to get involved.
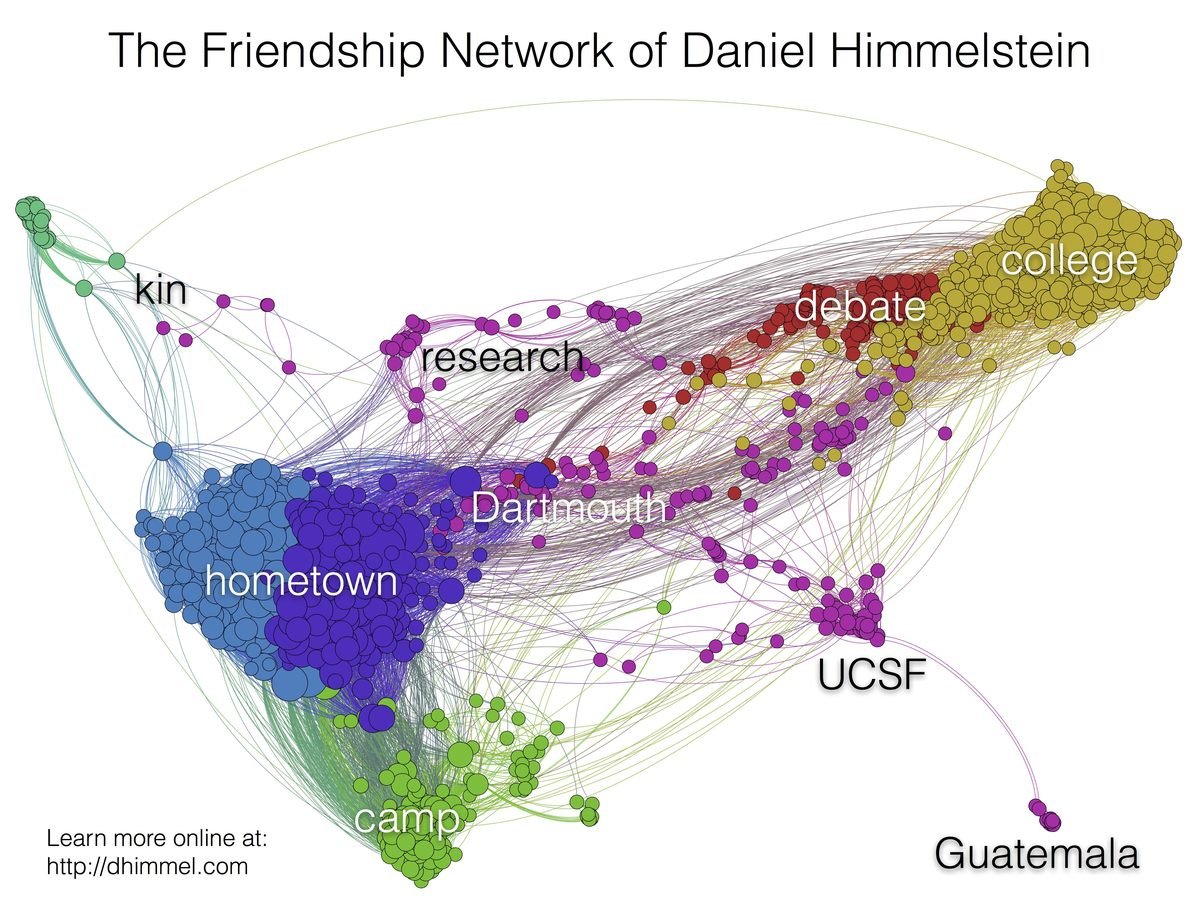
How I became intestested in graphs
http://blog.dhimmel.com/friendship-network/
The Graph Mindset
networks with multiple node or relationship types
multilayer network, multiplex network, multivariate network, multinetwork, multirelational network, multirelational data, multilayered network, multidimensional network, multislice network, multiplex of interdependent networks, hypernetwork, overlay network, composite network, multilevel network, multiweighted graph, heterogeneous network, multitype network, interconnected networks, interdependent networks, partially interdependent networks, network of networks, coupled networks, interconnecting networks, interacting networks, heterogenous information network
A 2012 Study identified 26 different names for this type of network:
hetnet

Graphs are composed of:
- Nodes
- Relationships
Nodes / relationships have type:
- node labels
(person, course, university) - relationship types
(lecturer, institution)

- first_name: Daniel
- last_name: Himmelstein
- twitter: @dhimmel
- SSN: 012-34-5678

- catalog: CIS550
- title: Database & Information Systems
- units: 5
- catalog: EPID600
- title: Data Science for Biomedical Informatics
- units: 1

- url: www.upenn.edu
- founded: 1740
- type: private
- league: ivy




- grade: A-
- grade: B
- grade: F

What can we do with this graph?
- Course statistics:
How many students are in CIS 550?
- Course recommendations:
What courses do other students in CIS 550 take?
- Course scheduling:
What room should a course be in so it's nearby other courses that its students are enrolled in?
How do you teach a computer biology?

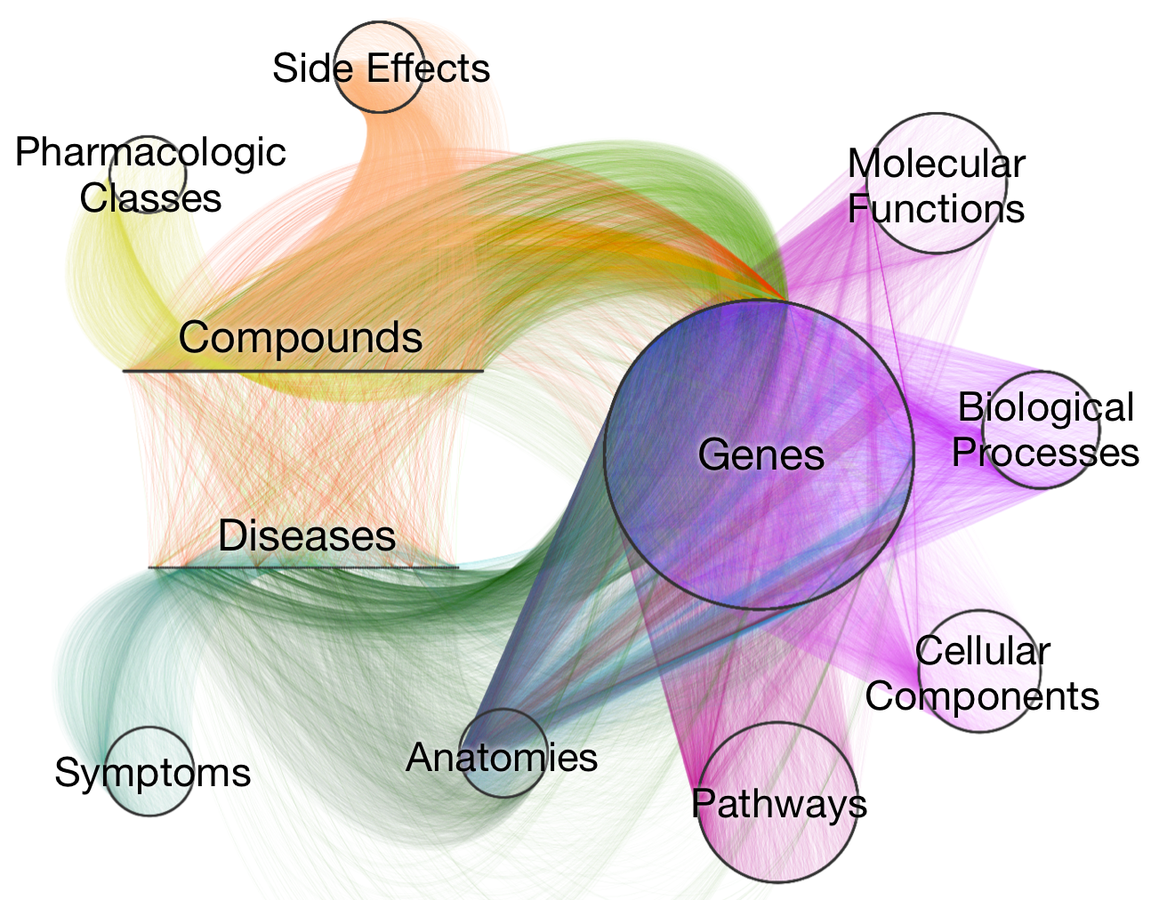
Visualizing Hetionet v1.0
- Hetnet of biology designed for drug repurposing
- ~50 thousand nodes
11 types (labels)
- ~2.25 million relationships
24 types
- integrates 29 public resources
knowledge from millions of studies
- the hardest part:
licensing of publicly available data
Hetionet v1.0
Nice of you to share this big network with everyone; however, I think you need to take care not to get yourself into legal trouble here. …
I am not trying to cause trouble here — just the contrary. When making a meta-resource, licenses and copyright law are not something you can afford to ignore. I regularly leave out certain data sources from my resources for legal reasons.
One network to rule them all
We have completed an initial version of our network. …
Network existence (SHA256 checksum for graph.json.gz) is proven in Bitcoin block 369,898.
-
Hetionet (≤ v1.0) integrated data from 31 resources:
- 5 United States Government works
- 12 openly licensed
- 4 non-commercial use only
- 9 were all rights reserved
- 1 explicitly & contractually forbid reuse
-
Requested permission for 11 resources:
- median time to first response was 16 days
- 2 affirmative responses
-
Other considerations:
- who owns data
- incompatibilities: share alike vs non-commercial
- copyright status of data & fair use
- Solution: license attribute per node/relationship
Legal barriers to data reuse
Recommendations:
- release data under an open license
- commit to open in your resource sharing plan
What's the best software for storing and querying hetnets?
| dhimmel/hetio | |
|---|---|
| 86 | |
| 5 | |
| 2 |



| neo4j/neo4j |
|---|
| 42,498 |
| 3,071 |
| 1,007 |
GitHub stats from 2016-10-09
- Customized Docker image
- Digital Ocean droplet
- SSL from Let's Encrypt
- readonly mode with a query execution timeout
- Custom GRASS style
- Custom guides
Public Hetionet Neo4j Instance
Details at doi.org/brsc
MATCH path =
// Specify the type of path to match
(n0:Disease)-[e1:ASSOCIATES_DaG]-(n1:Gene)-[:INTERACTS_GiG]-
(n2:Gene)-[:PARTICIPATES_GpBP]-(n3:BiologicalProcess)
WHERE
// Specify the source and target nodes
n0.name = 'multiple sclerosis' AND
n3.name = 'retina layer formation'
// Require GWAS support for the
// Disease-associates-Gene relationship
AND 'GWAS Catalog' in e1.sources
// Require the interacting gene to be
// upregulated in a relevant tissue
AND exists(
(n0)-[:LOCALIZES_DlA]-(:Anatomy)-[:UPREGULATES_AuG]-(n2))
RETURN pathHow could multiple sclerosis could affect retina layer formation?
More queries at thinklab.com/d/220
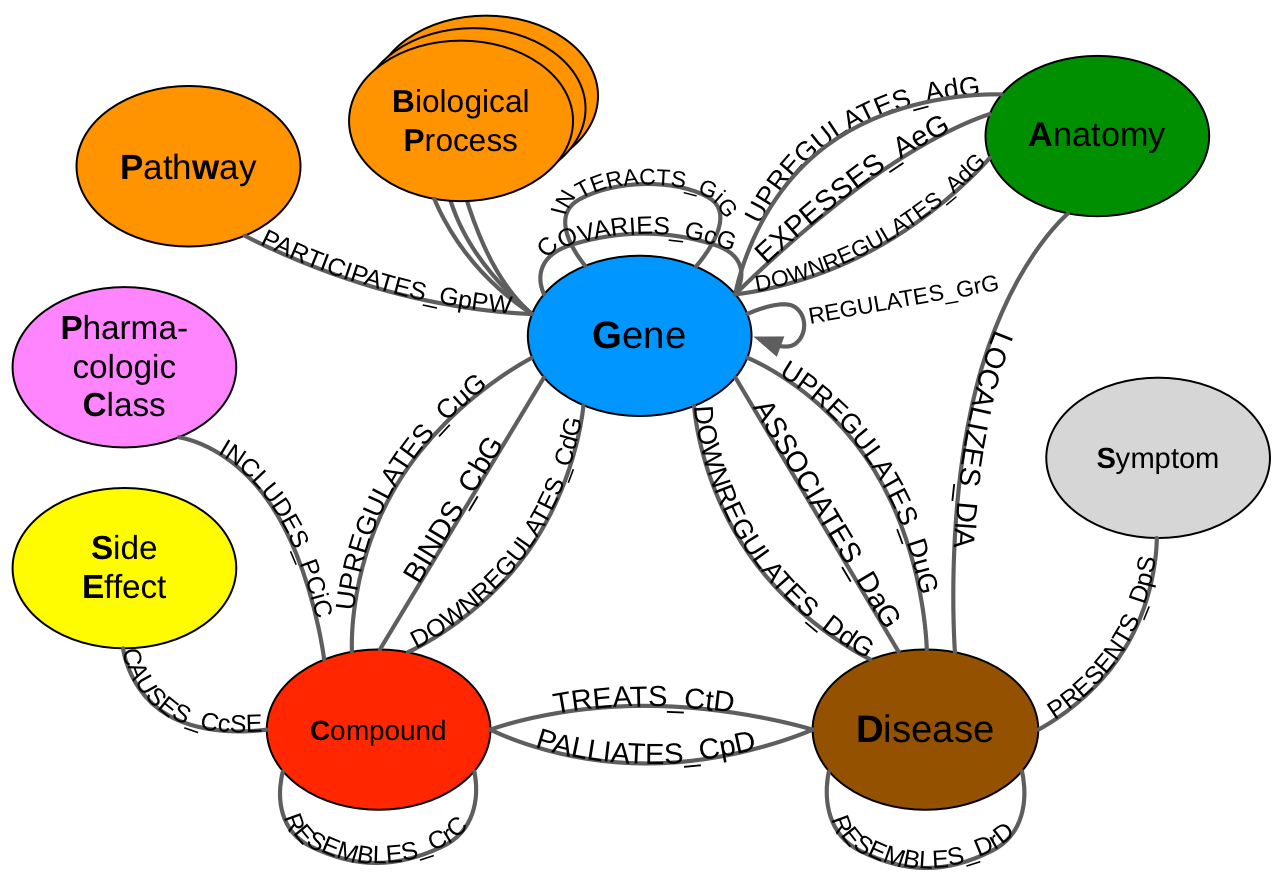
Neo4j Metagraph Nomenclature
Project Rephetio: drug repurposing predictions
-
Hetionet v1.0 contains:
- 1,538 connected compounds
- 136 connected diseases
- 209,168 compound–disease pairs
- 755 treatments
- Systematic drug repurposing:
- Compare the therapeutic utility of data types
- Identify the mechanisms of drug efficacy
- Predict the probability of treatment for all 209,168 compound–disease pairs (het.io/repurpose)
- Project online at thinklab.com/p/rephetio
Systematic integration of biomedical knowledge prioritizes drugs for repurposing
Daniel S Himmelstein, Antoine Lizee, Christine Hessler, Leo Brueggeman, Sabrina L Chen, Dexter Hadley, Ari Green, Pouya Khankhanian, Sergio E Baranzini
bioRxiv. 2016. DOI: 10.1101/087619

features = metapaths
observations =
compound–disease pairs
positives = treatments
negatives =
non-treatments
Machine learning methodology
1,206 compound–disease metapaths (length ≤ 4)
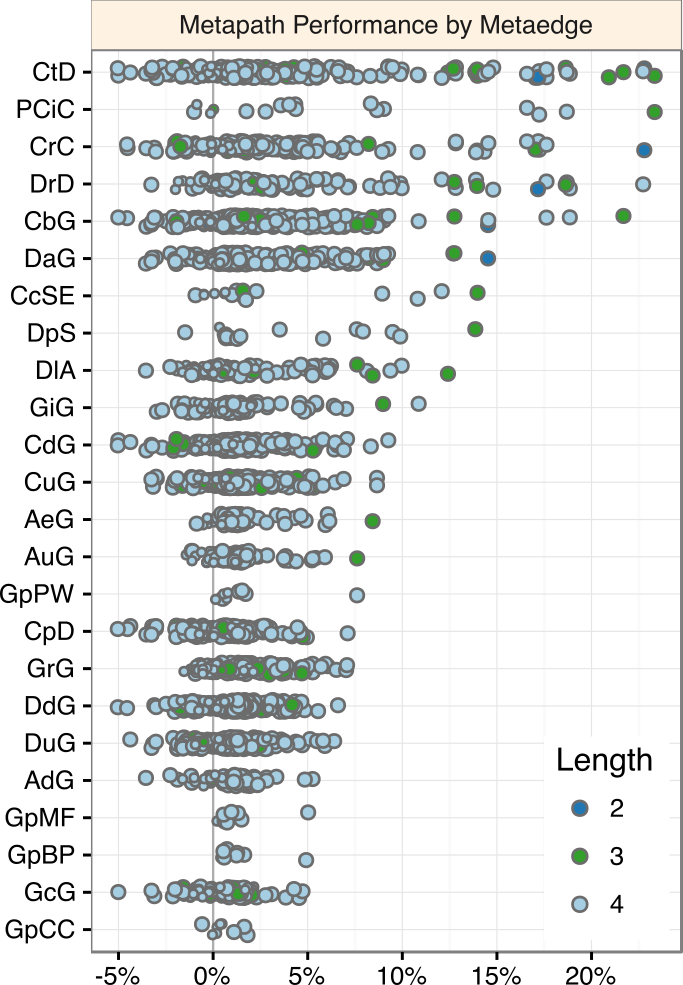
DWPC Δ AUROC
-
Upper tier:
traditional pharmacology -
Upper-middle tier:
traditionally biomedicine, but newer in drug efficacy -
Lower-middle tier:
genome-wide / high-throughput data sources -
Lower tier:
cellular components
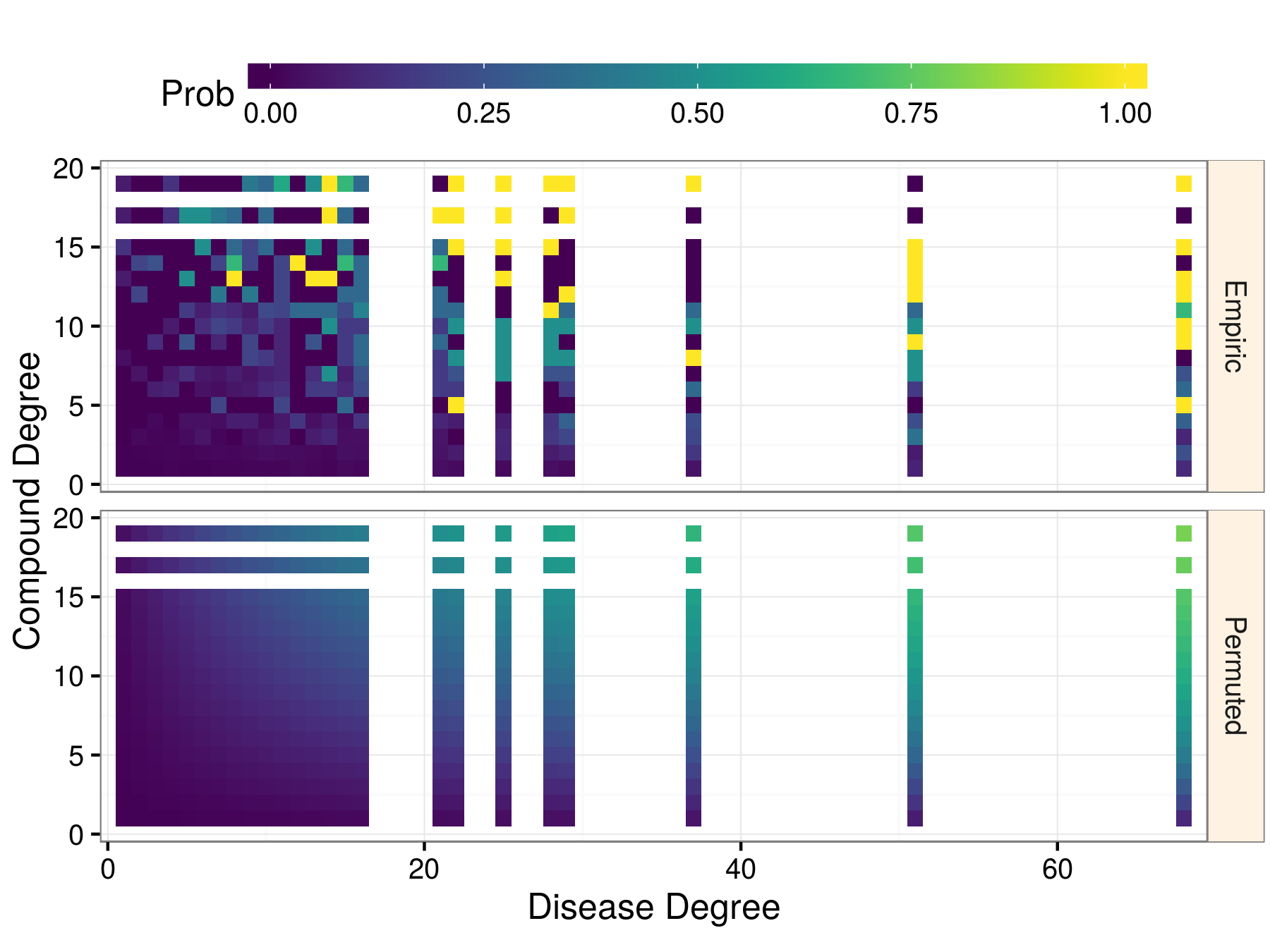
Prior probability of treatment
Methotrexate treats 19 diseases and hypertension is treated by 68 compounds. Methotrexate received a 79.6% prior probability of treating hypertension, whereas a compound and disease that both had only one treatment received a prior of 0.12%.
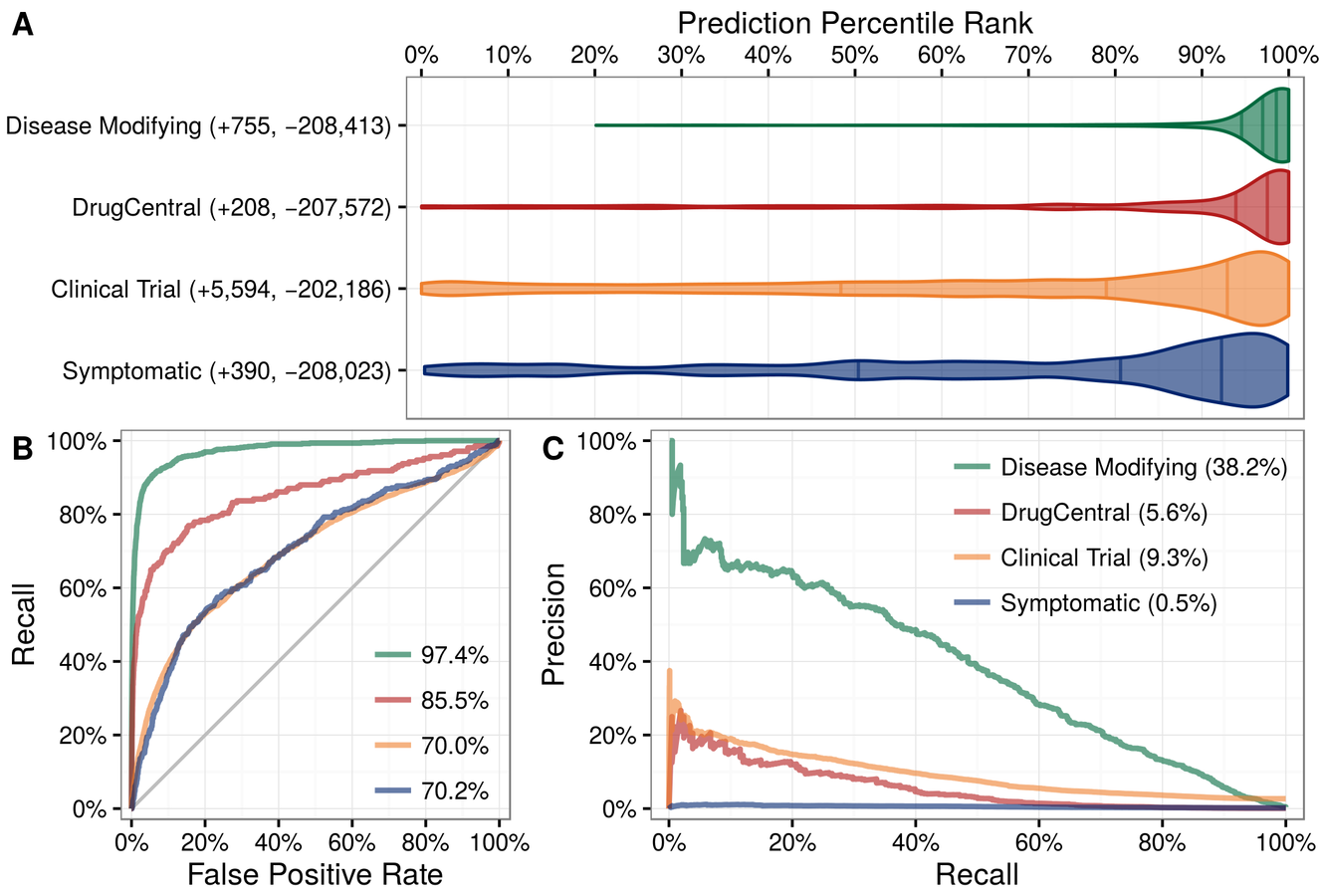
Predictions succeed at prioritizing known treatments

Project Rephetio: Does bupropion treat nicotine dependence?
- Bupropion was first approved for depression in 1985
-
In 1997, bupropion was approved for smoking cessation
- Can we predict this repurposing from Hetionet? The prediction was:
- 99.5th percentile for nicotine dependence
- probability 2.50-fold greater than null

Compound–causes–SideEffect–causes–Compound–treats–Disease

Compound–binds–Gene–binds–Compound–treats–Disease

Compound–binds–Gene–associates–Disease

Compound–binds–Gene–participates–Pathway–participates–Disease
MATCH path = (n0:Compound)-[:BINDS_CbG]-(n1)-[:PARTICIPATES_GpPW]-
(n2)-[:PARTICIPATES_GpPW]-(n3)-[:ASSOCIATES_DaG]-(n4:Disease)
USING JOIN ON n2
WHERE n0.name = 'Bupropion'
AND n4.name = 'nicotine dependence'
AND n1 <> n3
WITH
[
size((n0)-[:BINDS_CbG]-()),
size(()-[:BINDS_CbG]-(n1)),
size((n1)-[:PARTICIPATES_GpPW]-()),
size(()-[:PARTICIPATES_GpPW]-(n2)),
size((n2)-[:PARTICIPATES_GpPW]-()),
size(()-[:PARTICIPATES_GpPW]-(n3)),
size((n3)-[:ASSOCIATES_DaG]-()),
size(()-[:ASSOCIATES_DaG]-(n4))
] AS degrees, path
RETURN
path,
reduce(pdp = 1.0, d in degrees| pdp * d ^ -0.4) AS path_weight
ORDER BY path_weight DESC
LIMIT 10Cypher query to find the top CbGbPWaD paths
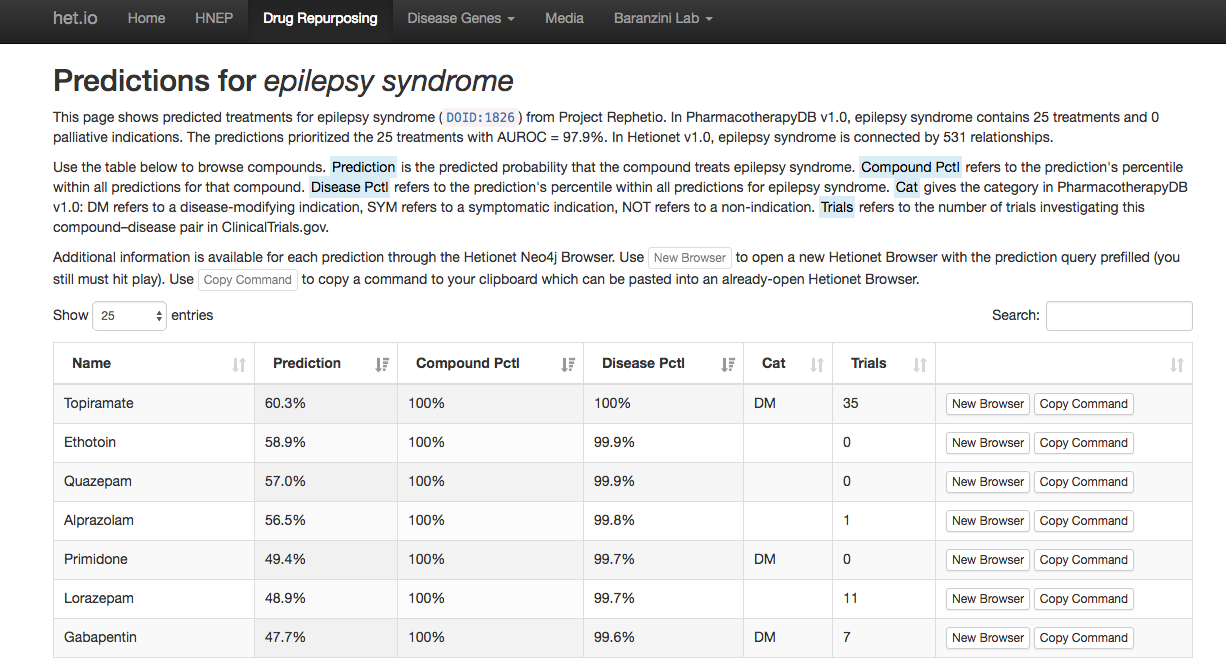
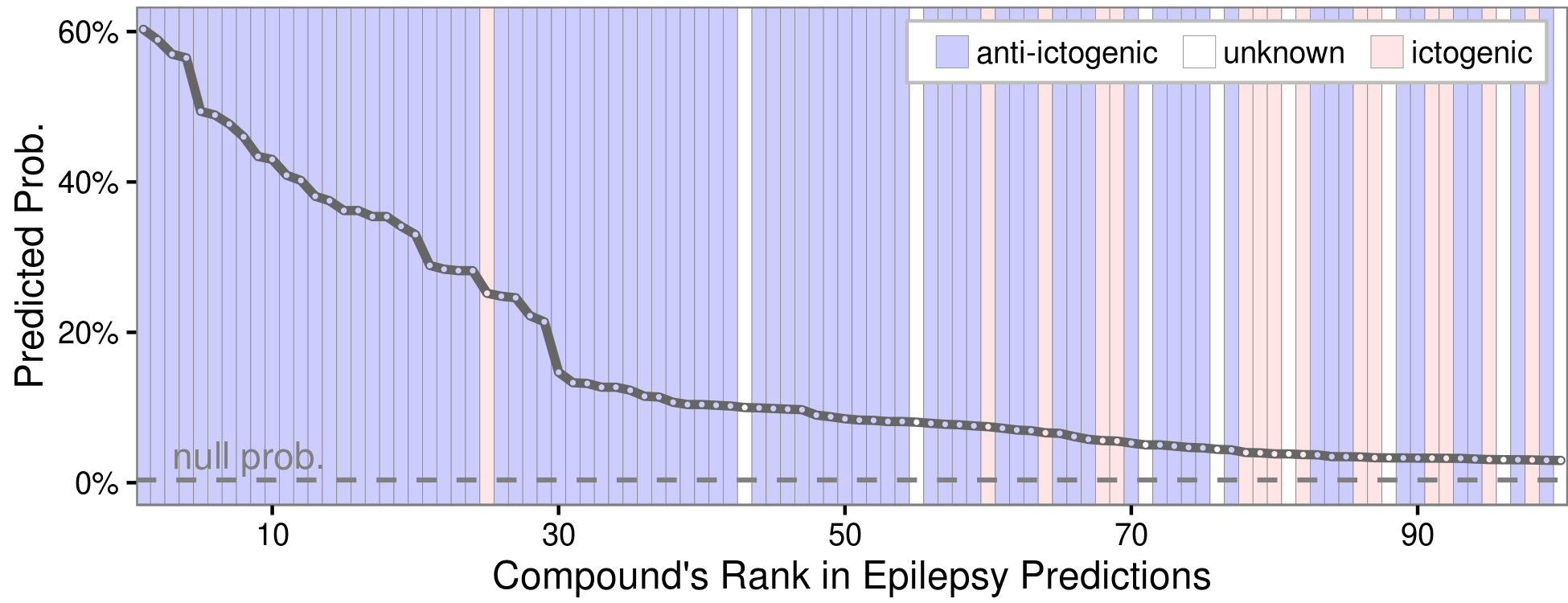
Epilepsy predictions
(browse all predictions at het.io/repurpose)
Discuss at thinklab.com/d/224
Evaluating the top 100 epilepsy predictions
Discuss at thinklab.com/d/224#5
Top 100 epilepsy predictions & their chemical structure
Discuss at thinklab.com/d/224#5
Top 100 epilepsy predictions & their drug targets
Discuss at thinklab.com/d/230#14
- 48 contributors
(see leaderboard) - 86 discussions
- 607 comments
- 143,056 words
- 901,672 characters
- ~20 publications
- https://thinklab.com/p/rephetio
Systematic integration of biomedical knowledge prioritizes drugs for repurposing
Daniel S Himmelstein, Antoine Lizee, Christine Hessler, Leo Brueggeman, Sabrina L Chen, Dexter Hadley, Ari Green, Pouya Khankhanian, Sergio E Baranzini
bioRxiv. 2016. DOI: 10.1101/087619
Project Rephetio contributions on Thinklab
(see thinklab.com/p/rephetio/leaderboard)
Future: all biomedical knowledge in a single network
https://github.com/greenelab/snorkeling

- Teach computers how to read the literature and extract knowledge.
- Continuously and automatically refine and grow the hetnet.
- Free from any legal restrictions on reuse.


Tissue-support requires both genes in the path to be expressed in the cardiovascular system.
Find the tissue-supported contribution of each pathway to treating CAD with enalapril (https://neo4j.het.io)
MATCH path = (n0:Compound)-[:BINDS_CbG]-(n1)-[:PARTICIPATES_GpPW]-
(n2)-[:PARTICIPATES_GpPW]-(n3)-[:ASSOCIATES_DaG]-(n4:Disease)
MATCH (n4)-[:LOCALIZES_DlA]-(anatomy)
MATCH (n1)-[:EXPRESSES_AeG]-(anatomy)-[:EXPRESSES_AeG]-(n3)
WHERE n0.name = 'Enalapril'
AND n4.name = 'coronary artery disease'
AND n1 <> n3
WITH
DISTINCT path,
n2 AS pathway,
[
size((n0)-[:BINDS_CbG]-()),
size(()-[:BINDS_CbG]-(n1)),
size((n1)-[:PARTICIPATES_GpPW]-()),
size(()-[:PARTICIPATES_GpPW]-(n2)),
size((n2)-[:PARTICIPATES_GpPW]-()),
size(()-[:PARTICIPATES_GpPW]-(n3)),
size((n3)-[:ASSOCIATES_DaG]-()),
size(()-[:ASSOCIATES_DaG]-(n4))
] AS degrees
RETURN
pathway.identifier AS pathway_id,
pathway.name AS pathway_name,
count(*) AS PC,
sum(reduce(pdp = 1.0, d in degrees| pdp * d ^ -0.4)) AS DWPC
ORDER BY DWPC DESC, pathway_nameQuestions
Slides at slides.com/dhimmel/mia