Hetnets & open science for healthcare: integrating biomedical knowledge to predict drug efficacy
Daniel Himmelstein (@dhimmel)
DeCART Summer Program
University of Utah, HSEB 2600
July 17, 2019 12:00 PM
slides released under CC BY 4.0
Encoding 50 years of biomedical knowledge into a network to predict when a drug treats a disease
Daniel Himmelstein (@dhimmel)
Huntsman Cancer Institute, University of Utah
HCI RS Conference Room 2C
July 16, 2019 12:00 PM
slides released under CC BY 4.0
Greene Lab
I'm a postdoc
http://www.greenelab.com/
Special thanks to
- Vince Rubinetti
- Michael Zietz
- Casey Greene
- Online collaborators
Short Abstract:
How can we encode all biomedical knowledge into a single resource optimized for machine learning? We explore using hetnets (networks with multiple node and relationship types) to integrate diverse information. By combining 29 public databases, we created Hetionet, a network with 11 node and 24 relationship types (available at https://neo4j.het.io). Next, we learned which types of paths occur more frequently when a drug treats a disease, allowing us to make over 200,000 predictions of treatment efficacy. Now we are creating a search engine at https://search.het.io/ to allow any researcher to quickly find how any two nodes in the hetnet are meaningfully connected. These studies were made possible by adopting a set of radically open practices, where all research was shared and discussed publicly from its inception. This includes our new Manubot software for open scholarly writing on GitHub.
Short Bio:
Daniel Himmelstein is a postdoctoral fellow in the Greene Lab at the University of Pennsylvania. Previously, he received his PhD from the University of California San Francisco. His research focuses on integrating biomedical knowledge using hetnets. Daniel is also a frequent contributor to open source/data ecosystems, and explores how computational research can become more open and reproducible.
DeCART Metadata
Hetionet
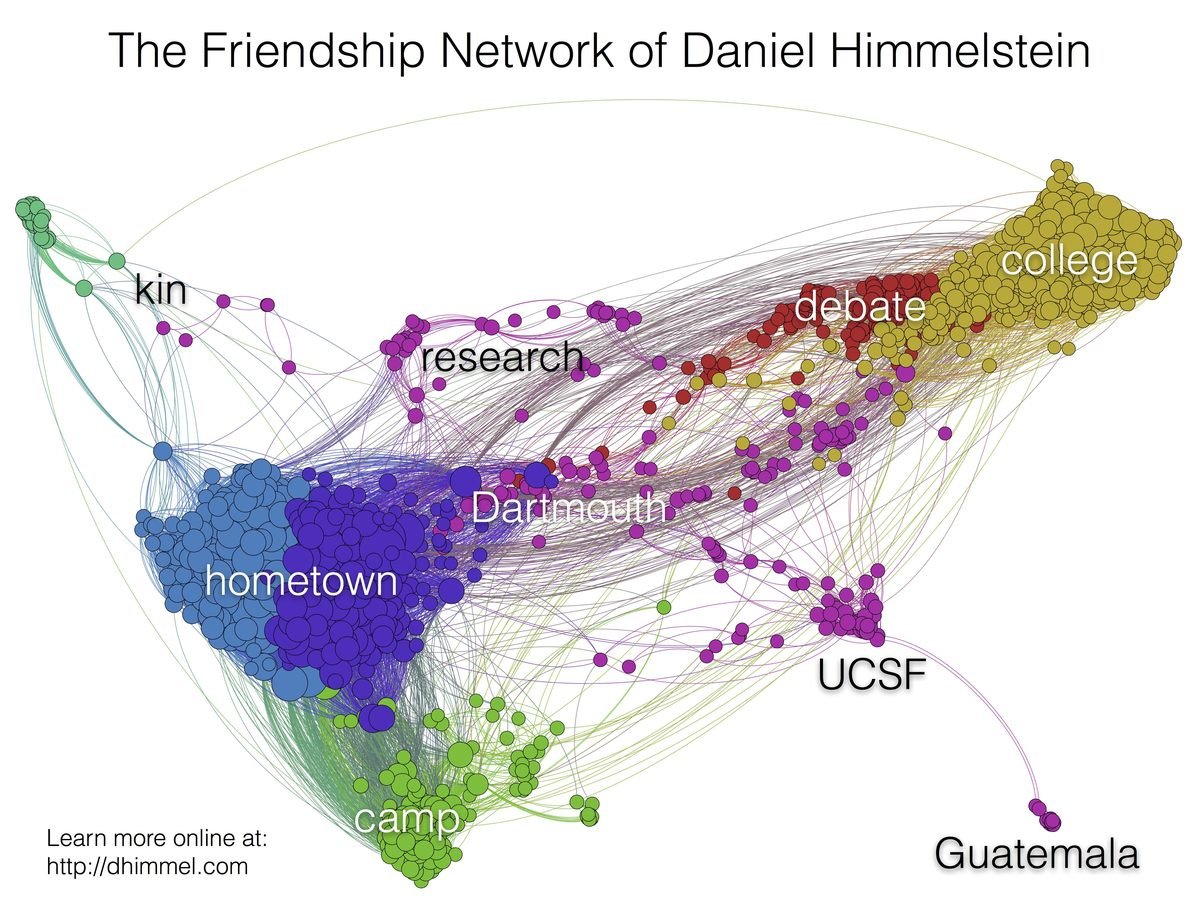
How I became intestested in graphs
http://blog.dhimmel.com/friendship-network/
My Facebook friendship network in 2014
too simple
single node type
single relationship type
networks with multiple node or relationship types
multilayer network, multiplex network, multivariate network, multinetwork, multirelational network, multirelational data, multilayered network, multidimensional network, multislice network, multiplex of interdependent networks, hypernetwork, overlay network, composite network, multilevel network, multiweighted graph, heterogeneous network, multitype network, interconnected networks, interdependent networks, partially interdependent networks, network of networks, coupled networks, interconnecting networks, interacting networks, heterogenous information network
A 2012 Study identified 26 different names for this type of network:
hetnet
How do you teach a computer biology?

online discussion contributions
(see thinklab.com/p/rephetio/leaderboard)
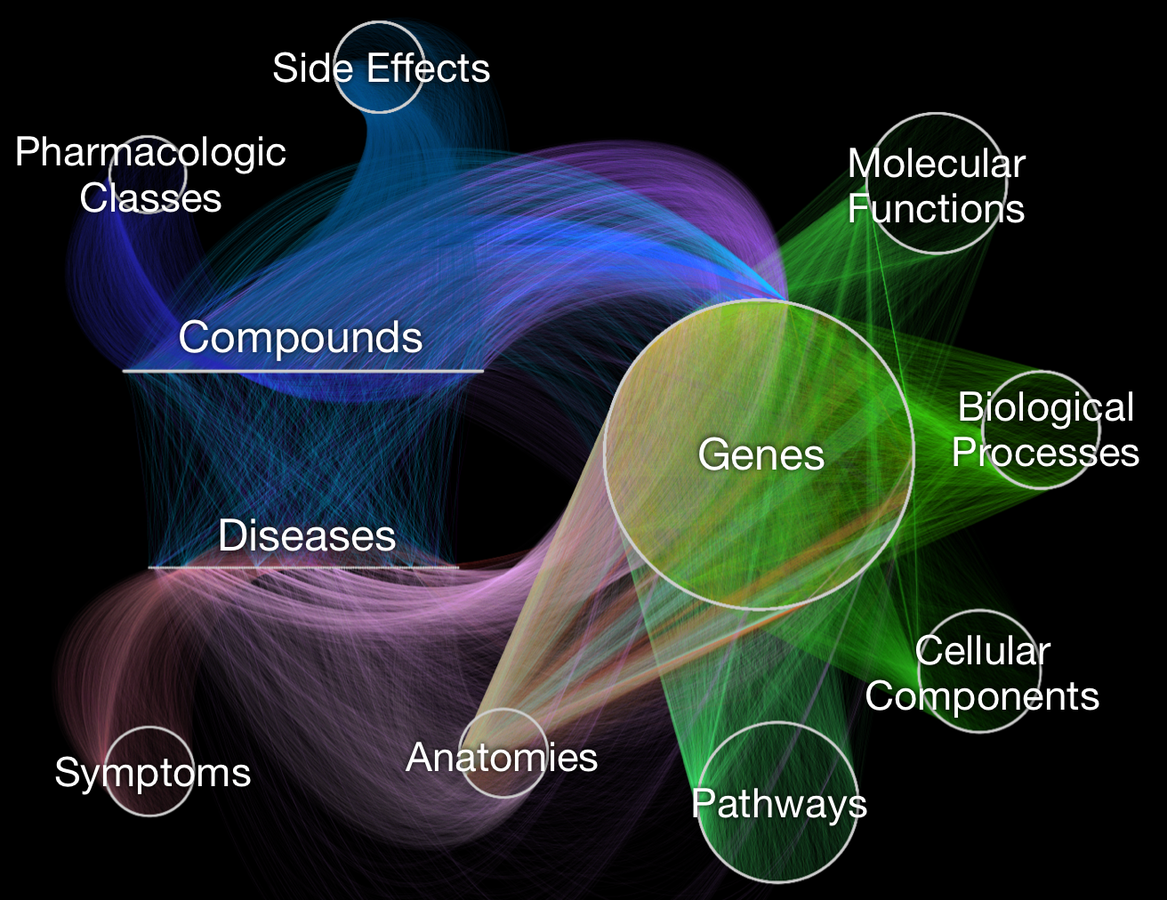
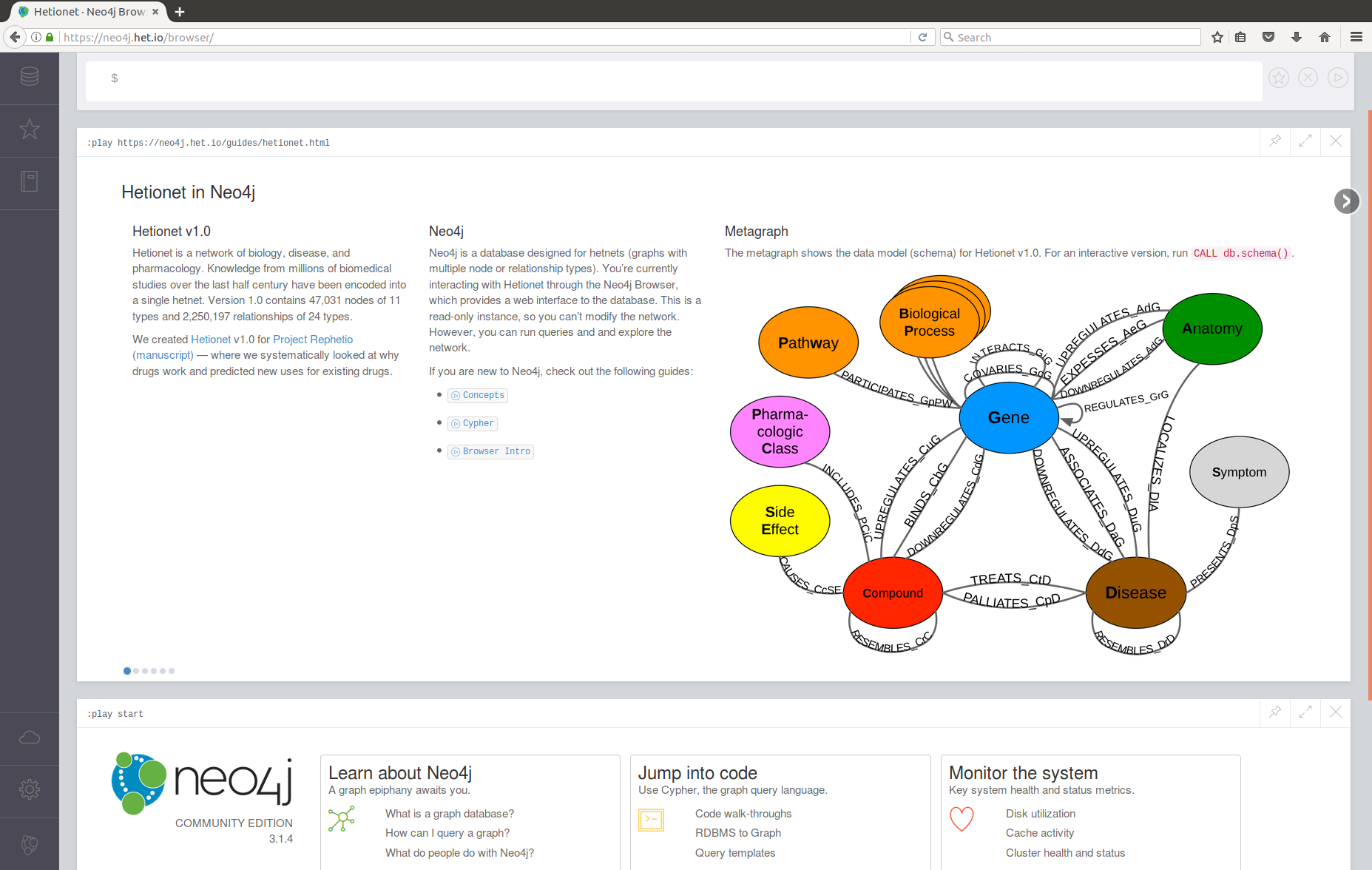
Visualizing Hetionet v1.0
- Hetnet of biology for drug repurposing
- ~50 thousand nodes
11 types (labels)
- ~2.25 million relationships
24 types
- integrates 29 public resources
knowledge from millions of studies
Hetionet v1.0
MATCH path =
// Specify the type of path to match
(n0:Disease)-[e1:ASSOCIATES_DaG]-(n1:Gene)-[:INTERACTS_GiG]-
(n2:Gene)-[:PARTICIPATES_GpBP]-(n3:BiologicalProcess)
WHERE
// Specify the source and target nodes
n0.name = 'multiple sclerosis' AND
n3.name = 'retina layer formation'
// Require GWAS support for the
// Disease-associates-Gene relationship
AND 'GWAS Catalog' in e1.sources
// Require the interacting gene to be
// upregulated in a relevant tissue
AND exists(
(n0)-[:LOCALIZES_DlA]-(:Anatomy)-[:UPREGULATES_AuG]-(n2))
RETURN pathHow could multiple sclerosis could affect retina layer formation?
More queries at thinklab.com/d/220
Rephetio
Project Rephetio: drug repurposing predictions
-
Hetionet v1.0 contains:
-
1,538 connected compounds
-
136 connected diseases
-
209,168 compound–disease pairs
-
755 treatments
-
-
Systematic drug repurposing:
-
Compare the therapeutic utility of data types
-
Identify the mechanisms of drug efficacy
-
Predict the probability of treatment for all 209,168 compound–disease pairs (het.io/repurpose)
-
Systematic integration of biomedical knowledge prioritizes drugs for repurposing
Daniel S Himmelstein, Antoine Lizee, Christine Hessler, Leo Brueggeman, Sabrina L Chen, Dexter Hadley, Ari Green, Pouya Khankhanian, Sergio E Baranzini
eLife (2017) https://doi.org/cdfk
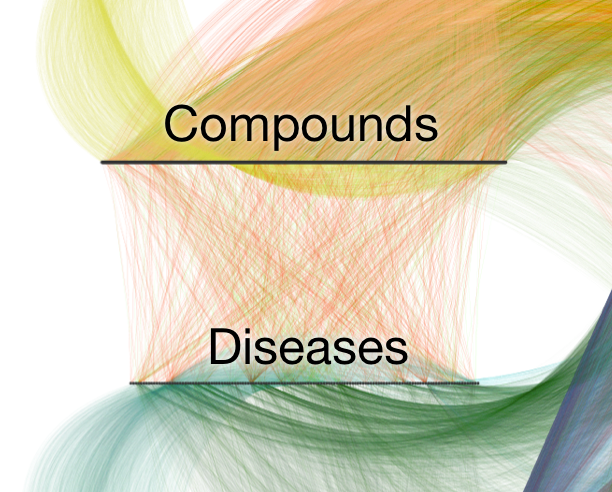

observations =
compound–disease pairs
features = types of paths
treatments
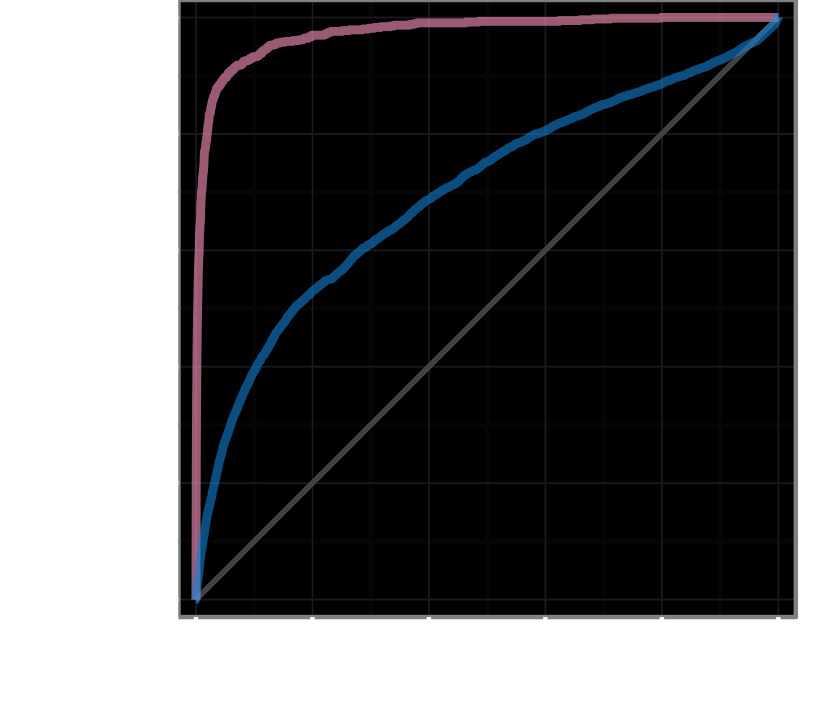
disease modifying treatments
+755, −208,413
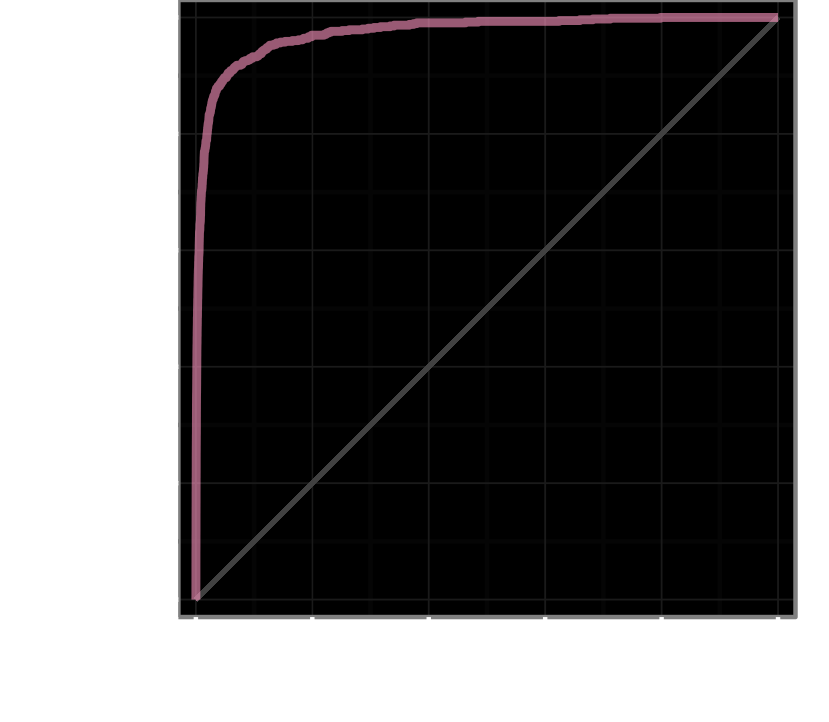
AUROC = 97.4%
treatments with clinical trials
+5,594, −202,186
AUROC = 70.0%
1,206 compound–disease metapaths (length ≤ 4)
Upper tier:
traditional pharmacology
Browse at het.io/repurpose/metapaths.html

Upper-middle tier:
traditionally biomedicine, but newer in drug efficacy
Lower-middle tier:
genome-wide / high-throughput data sources
Lower tier:
cellular components

Project Rephetio: Does bupropion treat nicotine dependence?
- Bupropion was first approved for depression in 1985
-
In 1997, bupropion was approved for smoking cessation
- Can we predict this repurposing from Hetionet? The prediction was:
- 99.5th percentile for nicotine dependence
- probability 2.50-fold greater than null

Compound–causes–Side Effect–causes–Compound–treats–Disease

Compound–binds–Gene–binds–Compound–treats–Disease

Compound–binds–Gene–associates–Disease

Compound–binds–Gene–participates–Pathway–participates–Disease
Extras
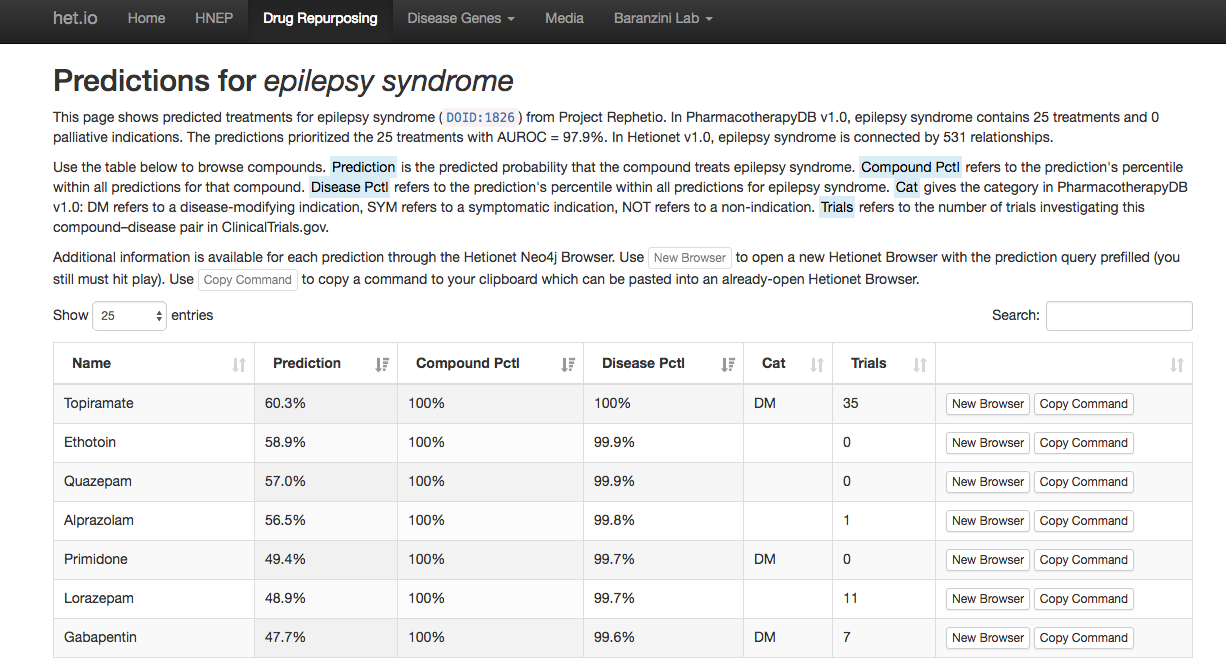
Browse all predictions at het.io/repurpose. Discuss at thinklab.com/d/224
Top 100 epilepsy predictions & their chemical structure
Discuss at thinklab.com/d/224#5
Top 100 epilepsy predictions & their drug targets
Discuss at thinklab.com/d/230#14
Connectivity Search
how are two nodes connected?
https://het.io/software/
https://het.io/search/
https://het.io/search/?source=17054&target=6602
findings → mechanims
we report that in human cancer cells, metformin inhibits mitochondrial complex I (NADH dehydrogenase) activity and cellular respiration.
— Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis
Wheaton et al (2014) eLife https://doi.org/gfpb2x
Metformin is the most widely used antidiabetic drug in the world, and there is increasing evidence of a potential efficacy of this agent as an anticancer drug. First, epidemiological studies show a decrease in cancer incidence in metformin-treated patients.
— Metformin in Cancer Therapy: A New Perspective for an Old Antidiabetic Drug?
Sahra et al (2010) Mol Cancer Ther https://doi.org/bgr5vv
Lung Cancer & Elevation
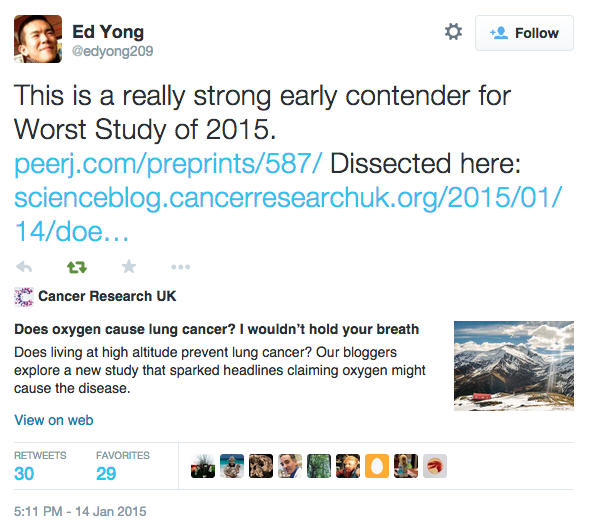
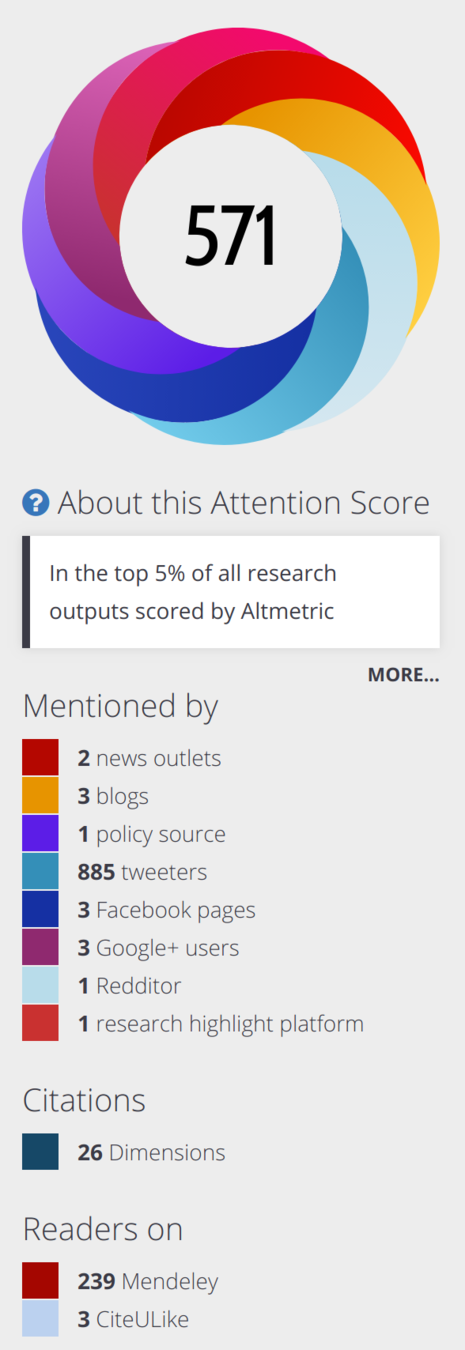
Simeonov & Himmelstein (2015) PeerJ. DOI: 10/98p
Salt Lake City: 1,288 meters
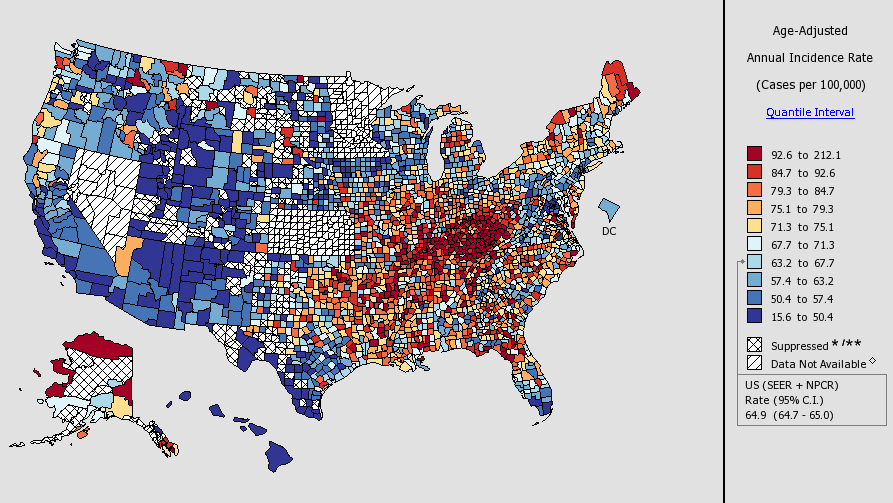
Lung Cancer Rates, 2007–2011
Source: NCI State Cancer Profiles


https://blog.dhimmel.com/elevation-and-lung-cancer/
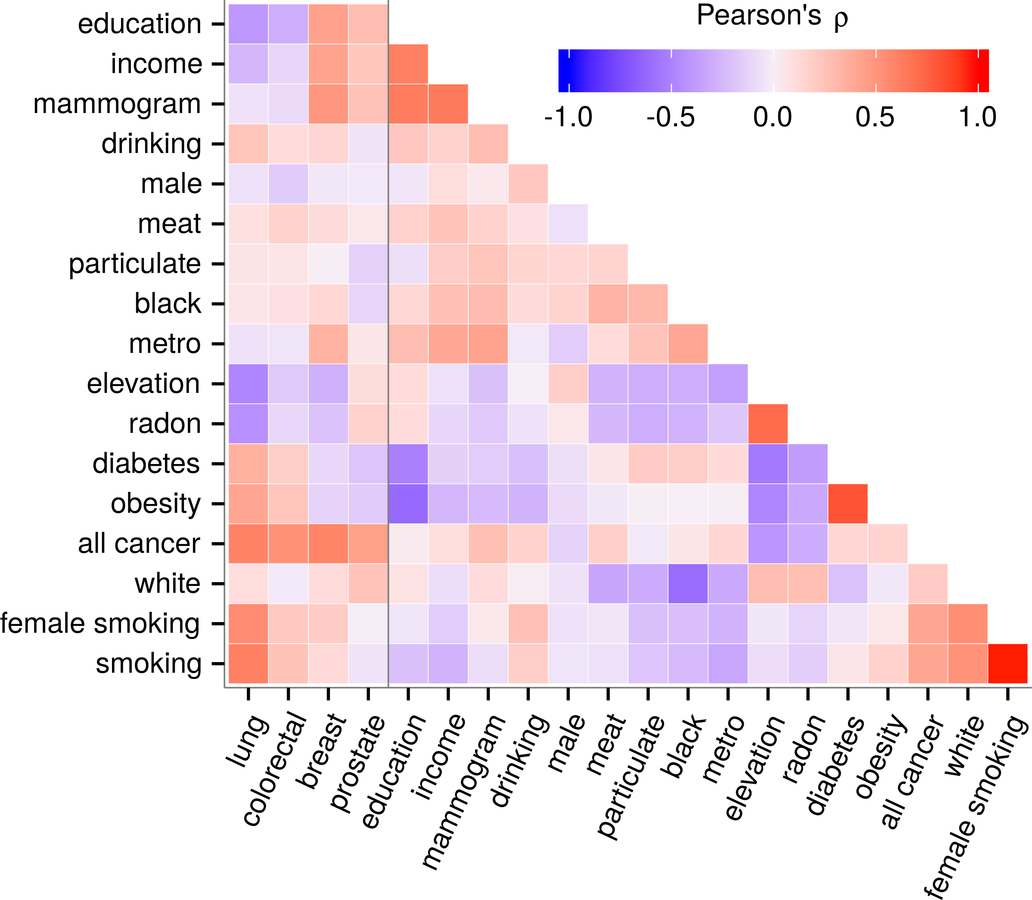
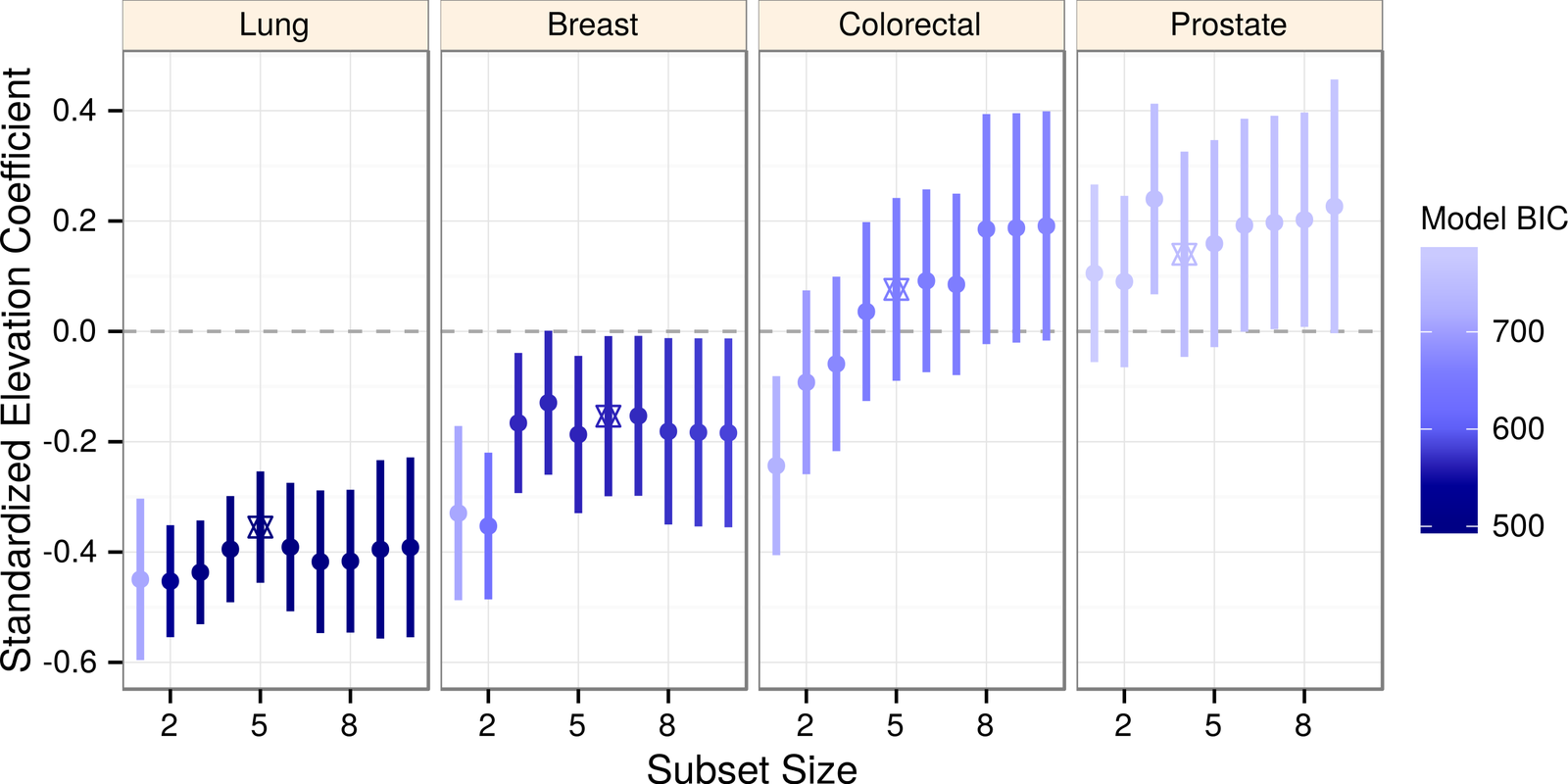
oxygen
relative to sea level:
-
89% at 1,000 meters
-
78% at 2,000 meters
-
69% at 3,000 meters
% tumor-free
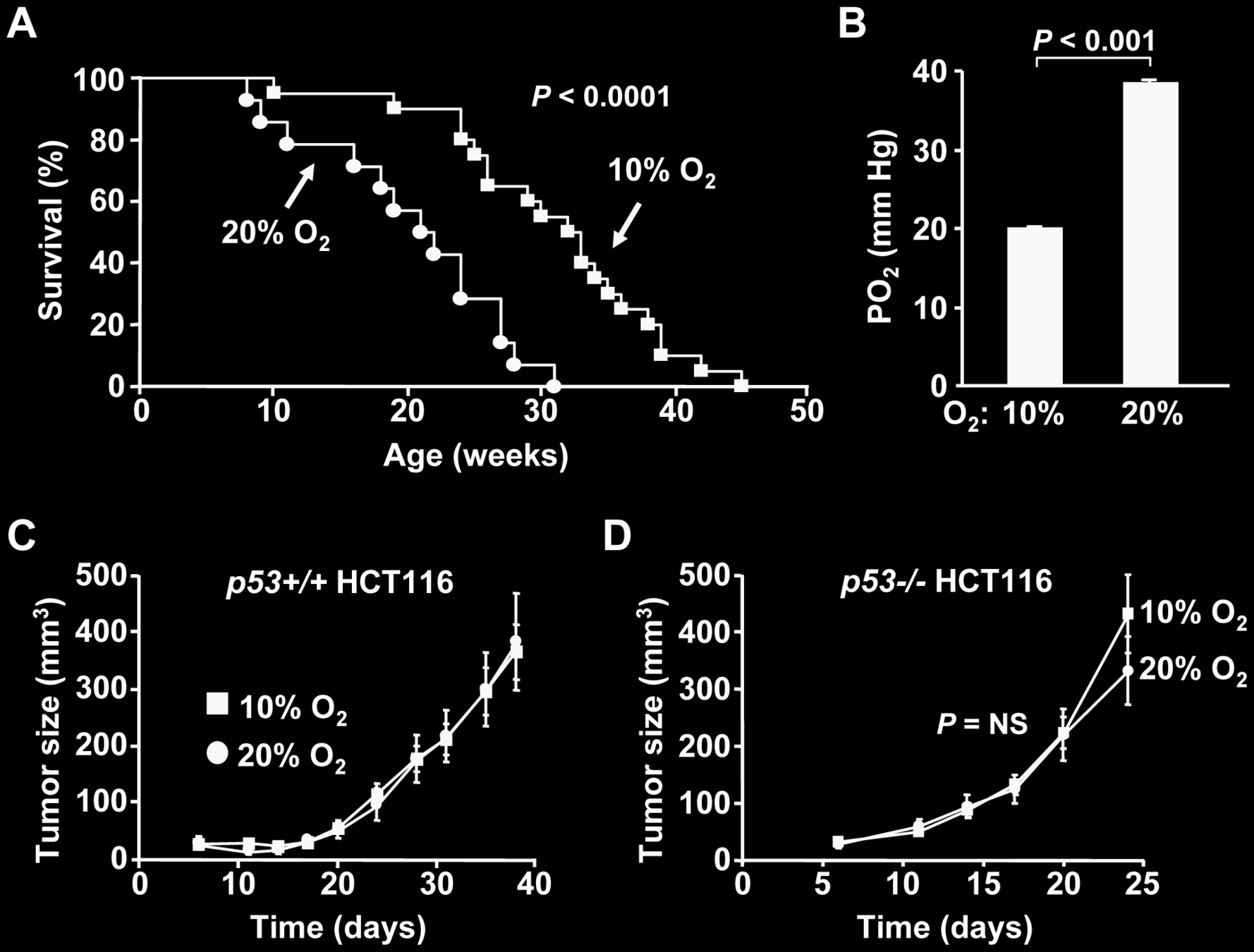
Ambient Oxygen Promotes Tumorigenesis
Sung & Ma et al (2011) PLOS One doi.org/fbcf2n
(in p53−/− mice)
Trouble at the Town Hall
1908 in Silverton, CO
San Juan Historical Society
San Juan County, Colorado
3,473 m
−35% O₂
−65,496


Our response is available at https://blog.dhimmel.com/cruk-reassessment/
2015 Abramson Cancer Center Basic Research Paper Prize
Unraveling the Ties of Altitude, Oxygen and Lung Cancer
George Johnson
Jon Krause



Manubot
Beyond the PDF First Day Notes
By De Jongens van de Tekeningen
Licensed under CC BY 3.0
Modified to invert colors
The Deep Review
- review article on deep learning in precision medicine
- 27 authors from 20 different institutions
- readers appreciate the breadth of perspectives
most viewed bioRxiv preprint of 2017
citation by persistent identifier



This is a sentence with 5 citations [ @doi:10.1038/nbt.3780; @pmid:29424689; @pmcid:PMC5938574; @arxiv:1407.3561; @url:https://greenelab.github.io/meta-review/ ].
References
-
Reproducibility of computational workflows is automated using continuous analysis
Brett K Beaulieu-Jones, Casey S Greene
Nature Biotechnology (2017-03-13) https://doi.org/f9ttx6
DOI: 10.1038/nbt.3780 · PMID: 28288103 · PMCID: PMC6103790
-
Sci-Hub provides access to nearly all scholarly literature.
Daniel S Himmelstein, Ariel Rodriguez Romero, Jacob G Levernier, Thomas Anthony Munro, Stephen Reid McLaughlin, Bastian Greshake Tzovaras, Casey S Greene
eLife (2018-03-01) https://www.ncbi.nlm.nih.gov/pubmed/29424689
DOI: 10.7554/elife.32822 · PMID: 29424689 · PMCID: PMC5832410
-
Opportunities and obstacles for deep learning in biology and medicine
Travers Ching, Daniel S. Himmelstein, Brett K. Beaulieu-Jones, Alexandr A. Kalinin, Brian T. Do, Gregory P. Way, Enrico Ferrero, Paul-Michael Agapow, Michael Zietz, Michael M. Hoffman, … Casey S. Greene
Journal of the Royal Society Interface (2018-04) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5938574/
DOI: 10.1098/rsif.2017.0387 · PMID: 29618526 · PMCID: PMC5938574
-
IPFS - Content Addressed, Versioned, P2P File System
Juan Benet
arXiv (2014-07-14) https://arxiv.org/abs/1407.3561v1
-
Open collaborative writing with Manubot
Daniel S. Himmelstein, David R. Slochower, Venkat S. Malladi, Casey S. Greene, Anthony Gitter
(2018-08-03) https://greenelab.github.io/meta-review/
This is a sentence with 5 citations [1,2,3,4,5].


Grant G-2018-11163 to DSH
https://manubot.org/catalog/
Future Directions
- learning from hetnets
- open source software for biomedical research
- tools to make science more open, transparent & reproducible
Thanks!
@dhimmel
0000-0002-3012-7446
Slides
https://slides.com/dhimmel/utah
Extra Slides


input
output
manubot process

Project Rephetio contributions on Thinklab
(see thinklab.com/p/rephetio/leaderboard)

- Nodes
- standardized vocabularies
- stable, unambiguous identifiers
- Relationships:
- Omics scale required
- Literature mining
- High throughput experimental technologies
- Avoid manual mapping
- Versioned data dependencies
Constructing Hetionet v1.0