




For Funding
Funder Requirements
- Ensure data is:
- accurate, complete
- authentic, reliable
- reusable
- Ensure:
- research integrity and reproducibility
- Minimizing risk of data loss
- Part of Open Research & Open data
A Datamanagement Plan, Why?

To avoid

For You
Improve research efficiency
- All time overview of data
- Better data management
- versioning
- avoid data duplication
- Better data protection
- Understand the data you created earlier
- Saving time (in the long run)
- Helps the data publication proces
- Meet funder requirements
- Better planning
- Guidance
- Get Credit
A Datamanagement Plan, Why?

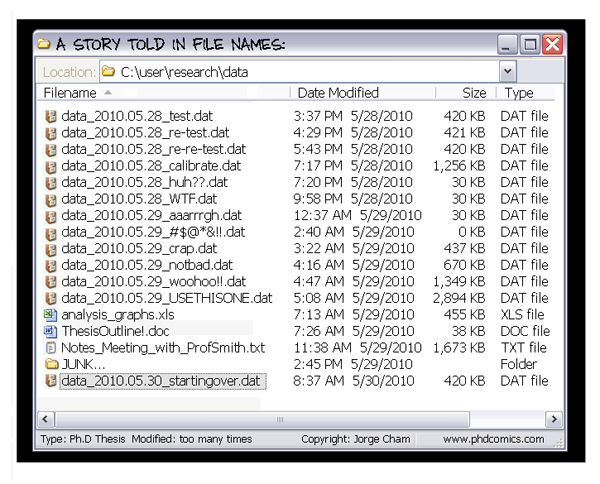
To avoid
For the Institute
- Part of the institutional data policy
A Datamanagement Plan, Why?

To avoid

For ME
A Datamanagement Plan, Why?

Improve research efficiency
- I can have an overview of your data
- I can understand your versioning
- I can understand your data
- Saving time (immediately)
- I can easily publish your data
- I can give you credit
My experience with DMP
or the non existence of the DMP

Under what licence shall we publish -->
- Go to the legal department
Can you send me your datasets -->
- ok, wait, I hope I can still find all the data
- ok, but I don't know where all the datasets are
- ok, but I need to clean the data first
- ok, but I first need to transform it to a readable format
- Oeps, I don't have it anymore
What will be the name of your dataset?
- huh?
My experience with DMP
or the non existence of the DMP

Do you have recorded metadata on the dataset -->
- huh? Metadata, what 's that?
What standard did you used?
- huh? a data standard?
Do your occurrences have global Unique ID's
- global unique what's?
something from my mailbox
Qx. hey, can you send me dataset X
Ay. ok, Dataset Y is doable, x is a mess, it s a mix betyween polygons and coördinates, maybe I can send you dataset Y

something from my mailbox
Qx. Ok, we could go for Y
Ay. Oh no wait, I first need to check this once agin

something from my mailbox
2 weeks later
Qx. Hey did you gave dataset Y some more thoughts
Ay. yes, only thoughts ;)
Ay. Hey I ve been thinking of you here is the data....

something from my mailbox
Ax: thanx alot
Qx: hey Y, I ve been checking your data, I can 't merge the table
Ay: Oeps, here we go again
Ay: I found it, I made a mistake, here is the next version
Ax: thanks a lot! here we go again

My experience with DMP
Having a DMP

Under what licence shall we publish -->
CC0 please
Can you send me your datasets -->
- ok, you can find them here
What will be the name of your dataset?
- Alien macroinvertebrates in Flanders
My experience with DMP
having a DMP

Do you have recorded metadata on the dataset -->
- Yes, you can find it here, it's in eml.
- http://www.gbif.org/dataset/3c428404-893c-44da-bb4a-6c19d8fb676a
What standard did you used?
- Everything is in DarwinCore
Do your occurrences have global Unique ID's
- Stupid question, off course they have
A Data Management Plan
for Biodiversity Research

Introduction
1. Project Title
Project ED/29 - “Possible Management Tools For Water Bodies In mega Landscapes (Panscape)”
2. Project Abstract
Ponds are still present in large numbers in various landscapes. They hold an important fraction of aquatic biodiversity, and include more rare species than any other non-marine aquatic habitat. The loss of small freshwater bodies will reduce connectivity among remaining populations, as well as reduce numbers of organisms (Gibbs, 1993; Semlitsch & Bodie, 1998). The Belspo project PANSCAPE, with partners in the Royal Belgian Institute of natural Sciences (Brussels) and the Universities of Ghent, Leuven, Liege and Namur, set out to develop integrated management tools for pools in various types of landscapes, namely: pristine, intermediate and extensive landuse (agricultural). Many groups of living organisms where monitored in 126 ponds, divided in clusters of three, each cluster representing the 3 types of land use. Both local and regional potential drivers of biodiversity were monitored. Presence/ absence of fish and macrophyte cover turned out to be sledgehammer drivers of aquatic biodiversity in ponds. Turbidity and sediment quality (sludge) are also important. The latter two are highly affected by presence of cattle (trampling). Regional drivers of biodiversity include land use. Effects are sometimes visible up to hundreds of meters. However, in general, local factors are more important drivers than regional ones. This will facilitate management in highly fragmented landscapes such as agricultural areas in Belgium. PANSCAPE confirms earlier results on high beta- and gamma-diversity amongst ponds. Older ponds might have higher biodiversity levels than new ponds. Therefore, optimal source allocation between creation of new ponds and maintenance of old ponds will be necessary. Also, old ponds might be source populations for new ponds. Incidence of parasites potentially harmful for cattle (example: liver fluke Fasciola hepatica) in the studied ponds in Belgium is very low, and farmers can thus be motivated to use ponds to water cattle, rather than use tapwater. Pesticide load is very low in the Belgian ponds, regardless of the land use, but heavy metals in areas under intensive anthropogenic activities should continue to be monitored.

Describe the data to be collected (actual observations) during your research including amount (if known) and format. Name the type of data, the instrument or collection approach, and how the data will be sampled. If actual data are interpreted, note the interpretation. Describe any quality control measures. Also describe the final derivative products (datasets and software or computer code) and the analysis used including analytical software packages that are required for replication, etc. Describe data (both digital and analog) and physical materials (samples and collections) gathered or generated during the time of the award.
Consider these questions:
- What data will be generated in the research?
- What data types (format) will you be creating or capturing? (e.g. experimental measures, qualitative, raw, processed)
- How will you capture or create the data? (This should cover content selection, instrumentation, technologies and approaches chosen, methods for naming, versioning, meeting user needs, etc, and should be sensitive to the location in which data capture is taking place)
- If you will be using existing data, state that fact and include where you got it. What is the relationship between the data you are collecting and the existing data?
- What language is the data?
Data Types, Format and Capture Methods

Data Types, Format and Capture Methods
This research project will collect a variety of data and samples from the fieldwork, as well as the laboratory analyses of those samples.
RAW data
Collection during 4 sampling events
Biological specimens (dataset, txt) --- Collection in the field --fieldnotes + specimens --> database --> files
Images of specimens (dataset, jpg) --- Collection in the field -- Camera --> files
Permanent collection abiotic variables
Waterlevel, pH, Temp datalogger (dataset, txt) --- permanent collection in the field -- VanEssen TD diver
Re-used data
Occurrence data on the same sampling locations downladed from GBIF (dataset, txt) --- www.gbif.org --> files (previously published under the BIOWINS project)
Preprocessed data
Genetic sequences (dataset, txt) --- In the lab -- PCR Analysis
Analyzed data
Data will be analyzed, visualized and processed with Matlab (dataset, .mat) and R packages(a,b,...) and Phyton packages (x,y,z)

The RAW data collected in the field and the occurrence data downloaded from GBIF will be imported in the Lab's biodiversity occurrences database . This database is capable of dealing with all the information that will be captured in the field, therefore we will not need to adapt the database datamodel. All the data in the database is secured and backed up automatically.
The Permanent collection abiotic variables will be stored as the original .txt files in the abiotic variables folder, gentic sequences will be kept in the PCR database under the Manscape project and will be extractded as .txt files in the genetic sequences folder. The DwC:occurrenceID will be used for each pcr proces.
Quality Control: automated validation of occurrence in the Lab's database and manual validation of flagged records by experts. All the scientific names will be compared and checked to the Institute Checklist. Abiotic data will be statistically checked for outliners.
All the datasets will be described and completed in English.
Data Types, Format and Capture Methods

Consider these questions:
- What form will the metadata describing/documenting your data take?
- How will you create or capture these details?
- Which metadata & data standards will you use and why have you chosen them? (e.g. accepted domain-local standards, widespread usage)
- What contextual details (metadata) are needed to make the data you capture or collect meaningful?
Standards and Metadata
Think about what details (metadata) someone else would need to be able to use these files. Describe the structural standards that you will apply in making data and metadata available. For example, for most ecological data, documentation should be structured in Ecological Metadata Language (EML). An example of metadata could also be as simple as a "readme file" to explain variables, structure of the files, etc.

The metadata for this project will be stored in .eml (ecological metadata standard), we will use the data management tool Morpho for this purpose. There will be 3 RAW datasets described, Occurrence Data, Abiotic Data, sequence Data. The metadatapackages will be saved in the respective dataset folder as a zip file. Digital photographs taken in the field will be labelled appropriately and saved in the respective samplingLocation folder. By querying the Occ Database we will extract te needed data from the lab's Occ database. These files will also be saved in the samplingLocation folder. The standard terms used for the occ data is the Darwin Core Standard (when fit). The DwC event Core structure will be used. gentic data will be saved to GeneBank. When possible, csv delimited plain text files will be used to store data, as they are easily imported to various analysis programs.
The whole folder structure and files will be versioned using GIT (Github PANSCAPE repository).
Standards and Metadata

Standards and Metadata
Folder Structure
Text
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\mappen.txt
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\OUTREACH
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\PROJECT
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 1
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 2
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 3
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 1\ABIOTIC DATA
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 1\OCC DATASET
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 1\PICTURES 1
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 1\SEQUENCE DATASET
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 2\ABIOTIC DATA
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 2\OCC data
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 2\PICTURES 2
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 2\SEQUENCe data
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 3\ABIOTIC DATA
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 3\OCC DATA
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 3\PICTURES
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\DATA\LOCATION 3\SEQUENCE DATA
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\PROJECT\Publications
C:\Users\dimitri_brosens\Documents\GitHub\PANSCAPE PROJECT\PROJECT\Publications\DRAFT
Explain how the responsibilities regarding the management of your data will be delegated. This should include time allocations, project management of technical aspects, training requirements, and contributions of non-project staff - individuals should be named where possible. Remember that those responsible for long-term decisions about your data will likely be the custodians of the repository/archive you choose to store your data. While the costs associated with your research (and the results of your research) must be specified in the Budget Justification portion of the proposal, you may want to reiterate who will be responsible for funding the management of your data.
Roles and Responsibilities
Consider the following:
- Outline the staff/organizational roles and responsibilities for implementing this data management plan.
- Who will be responsible for data management and for monitoring the data management plan?
- How will adherence to this data management plan be checked or demonstrated?
- What process is in place for transferring responsibility for the data?
- Who will have responsibility over time for decisions about the data once the original personnel are no longer available?

In this project we will partly use the Lab's occurrence database, curated by the Lab's DBA. The PI is responsible for organizing and labeling the imagery (.jpg). By using a github(shared) repository we will ensure that everybody will use the same folder structure and has access to all datasets there. After the project finishes the PI is responsible for the poblication on the data on the appropriate repositories.
Data Collection: Jan Jannssen (PI)
DBA (Database admin): Jim Hoos
Data validation responsible: TEAM Research Quality
DMP monitoring: Jan Jannssen (PI)
Data Publication: Jan Jannssen (PI)
Data Curation after project: TEAM open Data & Research
Roles and Responsibilities

Describe how and where you will make these data and metadata available to the community. Remember some funders are committed to timely and rapid data distribution; make sure you address how soon your data will be available. Indicate what data will be made available and preserved. Will data be accessible on a web page, by email request, via open-access repository, etc.?
Data Dissemination Methods
Consider these questions:
- What data will be made available from the study and preserved for the long-term?
- How and when will you make the data available? (Include resources needed to make the data available: equipment, systems, expertise, etc.)
- What transformations will be necessary to prepare data for preservation / data sharing?
- What metadata/ documentation will be submitted alongside the data or created on deposit/ transformation in order to make the data reusable?
- What related information will be deposited?
- What is the process for gaining access to the data?
- How long will the original data collector/creator/principal investigator retain the right to use the data before opening it up to wider use?
- Explain details of any embargo periods for political/commercial/patent or publisher reasons.

All the data collected and organized in this project will be published online in the appropriate repositories. This means that all the collected Occurrence data will be published to GBIF. The RAW data will be exported from the Lab's database to Darwin Core and will be published through the IPT instance of the Belgian Biodiversity Platform. The relevant images will also be be published by using the audubon Core. The images will be hosted on our fileserver. All the images will receive an uri.
The data used for analysis and the R code will be published on the github repository. This repository will be opened in the last semester of the project. The processed data, as in used for analysis and publications will be published on Dryad.
Data Dissemination Methods

Describe the policies under which these data will be made available. It is very important, the reason a DMP is required, that you specify how you will share your data with non-group members after the project is completed. If the data is of a sensitive nature—privacy or ecological endangerment concerns, for instance—and public access is inappropriate, address here the means by which granular control and access will be achieved (e.g. formal consent agreements, anonymized data, only available within a secure network, etc.).
Consider these questions:
- Will any permission restrictions need to be placed on the data?
- Are there ethical and privacy issues? If so, how will these be resolved?
- What have you done to comply with your obligations in your IRB Protocol?
- Who will hold the intellectual property rights to the data and how might this affect data access?
- What and who are the intended or foreseeable uses/users of the data?
- Do you plan on publishing findings which rely on the data? If so, do your prospective publishers place any restrictions on other avenues of publication?
Policies for Data Sharing and Public Access

All the raw data collected in this project will be published under a CC0 waiver. This means that the data will become public domain and there are no restrictions for the further use of this data. All the data will be become available in the last semester of the project. The github repository will also be open, so all the code generated or created in this project will be part of the public domain.
All data will be published under the FAIR principe:
- Findable
- Accessible
- Interoperable
- Re-usable
Policies for Data Sharing and Public Access

Policies for Data Sharing and Public Access
To be Findable:
• F1. (meta)data are assigned a globally unique and eternally persistent identifier.
• F2. data are described with rich metadata.
• F3. (meta)data are registered or indexed in a searchable resource.
• F4. metadata specify the data identifier.
To be Accessible:
• A1 (meta)data are retrievable by their identifier using a standardized communications protocol.
• A1.1 the protocol is open, free, and universally implementable.
• A1.2 the protocol allows for an authentication and authorization procedure, where necessary.
• A2 metadata are accessible, even when the data are no longer available.

To be Interoperable:
• I1. (meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation.
• I2. (meta)data use vocabularies that follow FAIR principles.
• I3. (meta)data include qualified references to other (meta)data.
To be Re-usable:
• R1. meta(data) have a plurality of accurate and relevant attributes.
• R1.1. (meta)data are released with a clear and accessible data usage license.
• R1.2. (meta)data are associated with their provenance.
• R1.3. (meta)data meet domain-relevant community standards.
Archiving, Storage and Preservation
Consider which data (or research products) will be deposited for long-term access and where. (What physical and/or cyber resources and facilities (including third party resources) will be used to store and preserve the data after the grant ends?)
Describe your long-term strategy for storing, archiving and preserving the data you will generate or use.
Text
Consider the following:
- What is the long-term strategy for maintaining, curating and archiving the data?
- Which archive/repository/database have you identified as a place to deposit data?
- What procedures does your intended long-term data storage facility have in place for preservation and backup?
- How long will/should data be kept beyond the life of the project?
- What data will be preserved for the long-term?
- On what basis will data be selected for long-term preservation?
- What metadata/documentation will be submitted alongside the data or created on deposit/transformation in order to make the data reusable?
- What related information will be deposited?

Archiving, Storage and Preservation
The data collected in the Panscape project will be archived as one .zip package in the Institutional data repository. This package will contain all the datasets and all the collected metadata. Further, all the datasets will be made available online by publishing them to the appropriate online data repositories as GBIF, Dryad & Genebank.
The metadata will be published as a datapaper, in a WoS indexed journal and will be linked to the different published datasets.

Long time storage:
| Data Type | Place | Standard | Licence |
|---|---|---|---|
| Occurrence | www.gbif.org | DwC | CC0 |
| Research Article | www.dryad.org | CC0 | |
| Code | www.github.com | CC0 | |
| Pictures | Fileserver | .jpg | CC-BY |
| Sequences | genbank.org | ASN.1 | CC0 |
| waterlevels | WaterMl2 | CC0 | |
Archiving, Storage and Preservation







http://www.gesis.org/fileadmin/upload/trainingcenter/images__documents/Research_data_management_questions.pdf

















