UNSUPERVISED FEATURE CONSTRUCTION AND KNOWLEDGE EXTRACTION FROM GENOME-WIDE ASSAYS OF BREAST CANCER WITH DENOISING AUTOENCODERS
Weiyuan
2018.02.01
Outline
- Research problem
- Background knowledge
- Key challenge
- Experiment
- Conclusion
Outline
- Research problem
- Background knowledge
- Key challenge
- Experiment
- Conclusion
Research problem
Emerging Techniques
- Modern genomic technologies enable people to acquire vast amount of data.
- Artificial Neural Networks (ANN) show new ways to do feature extraction.
Old methods
- Traditional methods are focused on known-unknowns, i.e. to discover a new disease-related gene. But not suitable for discovering new processes.
Emerging Techniques
Old methods
VS
Emerging Techniques
Old methods
VS
Apply ANN (dA) to large dataset providing new insights
Outline
- Research problem
- Background knowledge
- Key challenge
- Experiment
- Conclusion
Background Knowledge
Before we start ...
Autoencoder

Kudos to image author(s)
Autoencoder

Kudos to images author(s)
C
A
T
Autoencoder

Kudos to image author(s)
Nonlinearity
dA - Denoising Autoencoder

Kudos to image author(s)
dA - Denoising Autoencoder

Kudos to images author(s)



C
A
T

Outline
- Research problem
- Background knowledge
- Key challenge
- Experiment
- Conclusion
Key Challenge
Doctor: You need to take a pound of broccoli stalk because node 64726 in our network said so.
Doctor: You need to take a pound of broccoli stalk because node 64726 in our network said so.
Something goes wrong!
Interpretation is much important
- Linking features to sample characteristics
- Linking feature to transcription factors
- Linking feature to patient survival*
- Linking feature to biological pathways
Interpretation is much important
- Linking features to sample characteristics
Sample Characteristics
- ER status (ER: estrogen receptor. Estrogen is a specific hormone from females)
- Molecular subtypes
- ...
How
- For all features \(f_i\) from the output of node \(n_i\), get its output range \([lower_i, upper_i]\) on training dataset.
How
- For all features \(f_i\) from the output of node \(n_i\), get its output range \([lower_i, upper_i]\) on training dataset.
- Split range \([lower_i, upper_i]\) into 10 equally spaced threshold \(T_i = \{t_i^1, t_i^2, ..., t_i^{10} \}\).
How
- For all features \(f_i\) from the output of node \(n_i\), get its output range \([lower_i, upper_i]\) on training dataset.
- Split range \([lower_i, upper_i]\) into 10 equally spaced threshold \(T_i = \{t_i^1, t_i^2, ..., t_i^{10} \}\).
- Binarize the feature \(f_i\) into \(\{true, false\}\) for each \(t_i \in T_i\).
How
- For all features \(f_i\) from the output of node \(n_i\), get its output range \([lower_i, upper_i]\) on training dataset.
- Split range \([lower_i, upper_i]\) into 10 equally spaced threshold \(T_i = \{t_i^1, t_i^2, ..., t_i^{10} \}\).
- Binarize the feature \(f_i\) into \(\{true, false\}\) for each \(t_i \in T_i\).
- Select these features have most predictive power toward the target characteristic.
Interpretation is much important
- Linking features to sample characteristics
- Linking feature to transcription factors
- Linking feature to patient survival
- Linking feature to biological pathways
Interpretation is much important
- Linking feature to patient survival

The output features behave bimodally

Cut off at 0.5 for each feature

Binarize the feature, evaluate the predictive power toward patient survival
Interpretation is much important
- Linking features to sample characteristics
- Linking feature to transcription factors
- Linking feature to patient survival
- Linking feature to biological pathways
Outline
- Research problem
- Background knowledge
- Key challenge
- Experiment
- Conclusion
Datasets
- METABRIC (2/3) - discovery: \(2136 \times 2520\)
- METABRIC (1/3) - validation: \(2136 \times 2520\)
- TCGA - evaluation: \(547 \times 2520\)
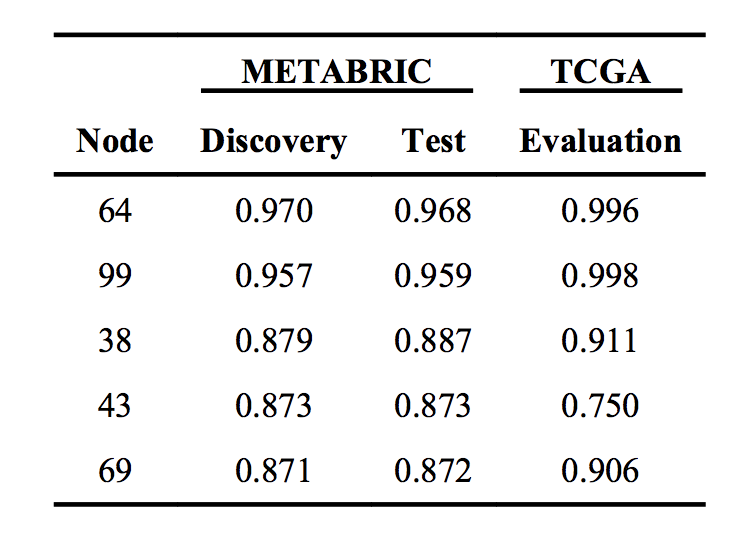
Performance of hidden nodes in classifying tumor from normal samples.

Performance of hidden nodes in classifying ER + from ER - samples.
Outline
- Research problem
- Background knowledge
- Key challenge
- Experiment
- Conclusion
Conclusion
- Features automatically generated by dA has good predictive power
- Using unsupervised model can fill the gap of the current method.
- Future work may involve supervised learning on these auto-generated features to increase accuracy.