Neuro Evolution Of Augmenting Topologies
@eliaswalyba
Writing Hieroglyphs. Building Pyramids.
GalsenAI Meetup
Oct. 5th, 2019
Agenda
- Introduction
- Understanding the NEAT Algorithm
- Discussing the NEAT Algorithm
- NEAT in Action
- Conclusion




P = NP ?
Nature may have the solution ?

In computer science, evolutionary computation is a family of algorithms for global optimization inspired by biological evolution, and the subfield of artificial intelligence and soft computing studying these algorithms.
Trial and error problem solvers.

Owkay nigga. Wait a minute Imma tweet this shit !
Problems with neural networks
- How many neurons / layer ?
- How many layers ?
- How many connections ?
- How to chose initial weights ?
- How to chose activation functions ?

Nigga listen, I am sure nature have the solution
1993
Let's remove backpropagation and gradient descent and evolve neural networks
2001
NEAT
Kenneth O. Stanley
@kenneth0stanley
Founding member, Uber AI Labs
UCF Prof. AI ML Neuroevolution: NEAT, HyperNEAT, novelty search.
Book: Why Greatness Cannot Be Planned

Instead of relying on a fixed structure for a neural network, why not allow it to evolve through a genetic algorithm?
- Is there a genetic representation that allows topologies to cross-over in a meaningful way ?
- How can a topological innovation that needs a few generations to be optimized be protected so that it doesn't disappear from the population prematurely ?
- How can topologies be minimized throughout evolution without the need for a specially contrivied fitness function that measures complexity ?
NEAT focuses solely on evolving dense neural networks node by node and connection by connection
Genotype
A genotype is the genetic representation of a creature
Phenotype
A phenotype is the actualized physical representation of the creature.
1
2
3
4
5
6
1
2
3
4
5
6
Phenotype
Genotype
| Node 1 sensor |
Node 2 Sensor |
Node 3 Sensor |
Node 4 Output |
Node 5 Output |
Node 6 Hidden |
| In: 1 Out: 5 Weight: 0.5 Enabled Innov 1 |
In: 2 Out: 5 Weight: 0.7 Disabled Innov 2 |
In: 2 Out: 6 Weight: 0.4 Enabled Innov 3 |
In: 3 Out: 6 Weight: 0.8 Enabled Innov 4 |
In: 6 Out: 4 Weight: 0.2 Enabled Innov 5 |
Input and output nodes are not evolved in the node gene list. Hidden nodes can be added or removed. As for connection nodes, they specify where a connection comes into and out of, the weight of such connection, whether or not the connection is enabled, and an innovation number
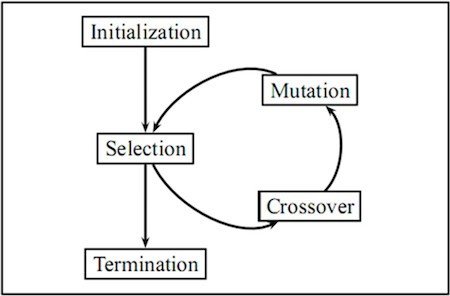
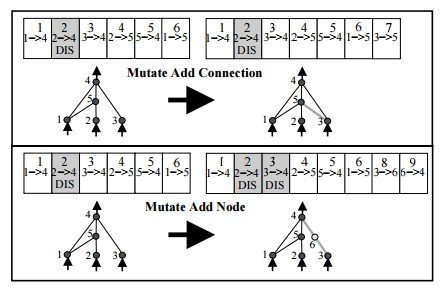
Mutation Golden Rules
- Mutate existing connections
- Add new structure to a network
- If a new connection is added between a start and end node, it is randomly assigned a weight.
- If a new node is added
- it is placed between two nodes that are already connected.
- The previous connection is disabled
- The previous start node is linked to the new node with the weight of the old connection
- The new node is linked to the previous end node with a weight of 1
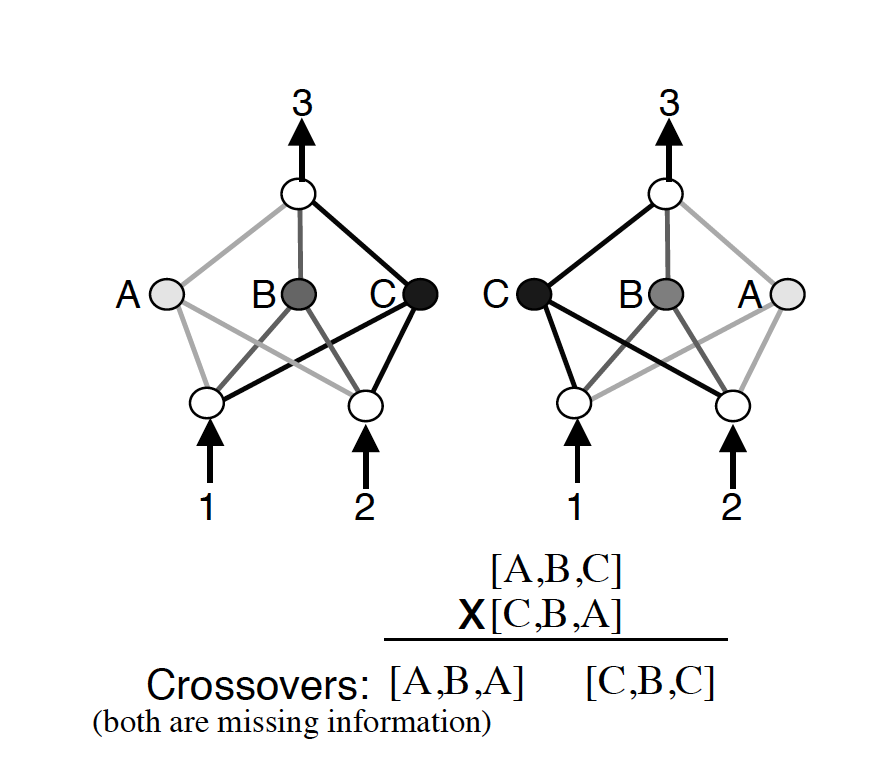
Another big issue with evolving the topologies of neural networks is something that the NEAT paper calls “competing conventions.” The idea is that just blindly crossing over the genomes of two neural networks could result in networks that are horribly mutated and non-functional. If two networks are dependent on central nodes that both get recombined out of the network, we have an issue.
Competing Convention
More than that, genomes can be of different sizes. How do we align genomes that don’t seem to be obviously compatible ?

Nigga don't worry ! Nature will solve your problem
In biology, this is taken care of through an idea called homology. Homology is the alignment of chromosomes based on matching genes for a specific trait. Once that happens, crossover can happen with much less chance of error than if chromosomes were blindly mixed together.
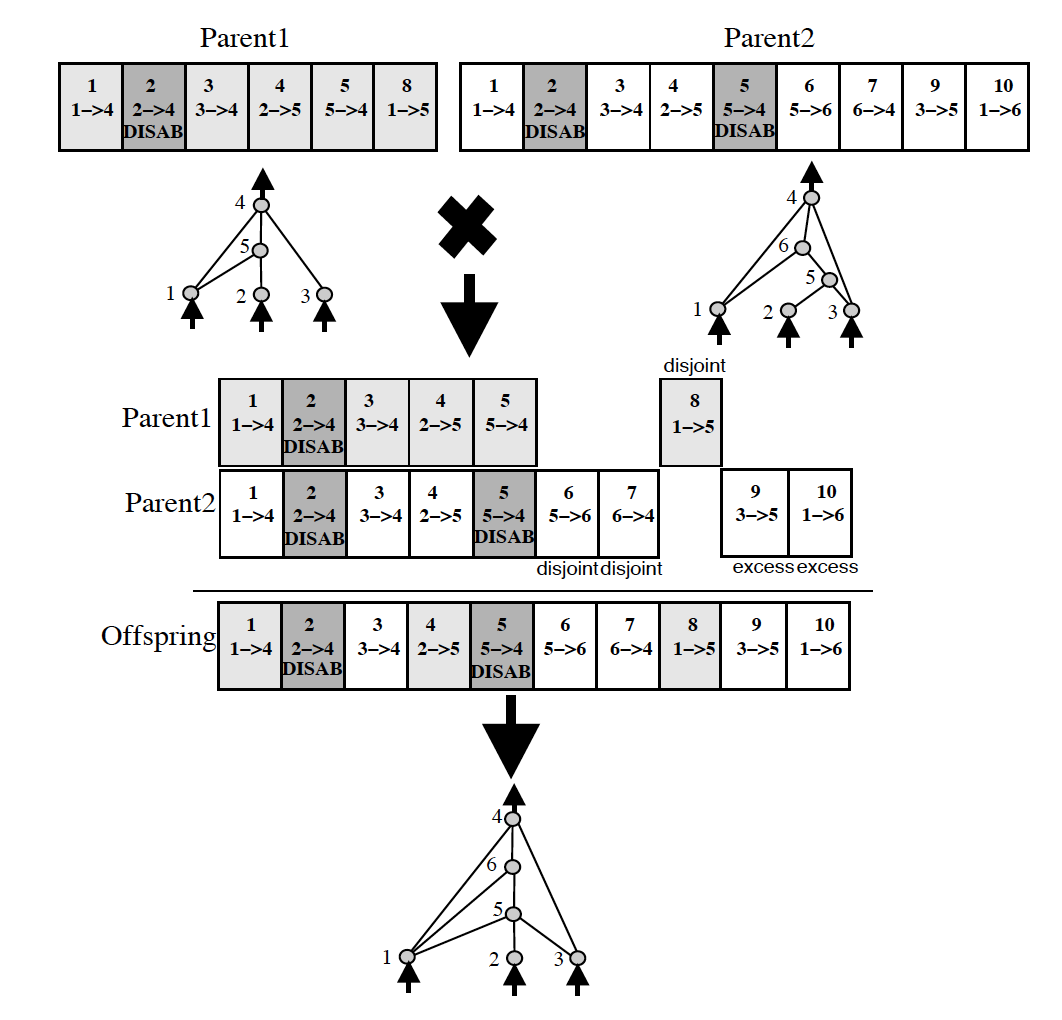
How can we protect new structures and allow them to optimize before we eliminate them from the population entirely ?
NEAT suggests speciation.
Speciation simply splits up the population into several species based on the similarity of topology and connections. If the competing convention problem still existed, this would be very hard to measure! However, since NEAT uses historical markings in its encoding, this becomes much easier to measure.
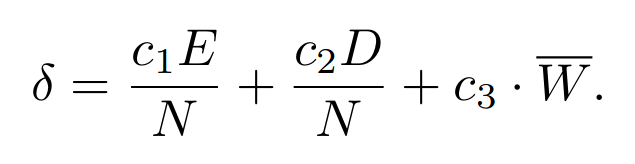
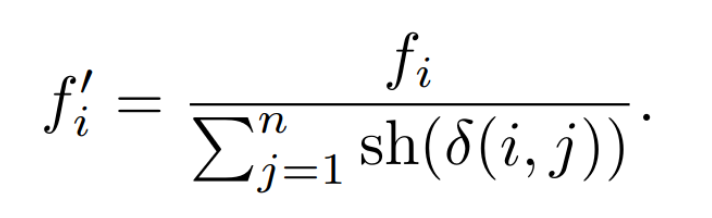
More than that, NEAT takes things one step forward through something called explicit fitness sharing. That means that individuals share how well they are doing across the species, boosting up higher performing species, though still allowing for other species to explore their structure optimization before being out evolved.

Hyper Parameter Optimization
- c1, c2, c3 fitness sharing parameters
- Initial population size
- Mutation Sampling Distribution
Is it useful to have an evolutionary algorithm optimizing an evolutionary algorithm optimizing a neural network ?
NEAT doesn't replace necessarily
Back Propagation + Gradient Descent