Data science tools
for reproducible neuroscience

live follow along: https://slides.com/ericearl/abcdworkshop1/live
or browse yourself: https://slides.com/ericearl/abcdworkshop1
Navigation
(look in the bottom right corner)

Previous topic
Next topic
Out of a topic
Into a topic
Take-aways
by the end of this talk, you should be able to say you can...
-
Find, browse, and clone GitHub repositories
-
Open a JupyterLab Notebook and use it
-
Run a Docker image (or a BIDS App)
-
Navigate BIDS data and metadata
-
Know where to find ABCD Study data
Prerequisite knowledge
You can open a terminal and run commands.
if terminals make you uncomfortable, read the below chapter:
Conquering the Command Line, Chapter 1: Basics and Navigation
Repeatable & Reproducible
repeatable
same team & same experimental setup
reproducible methods
different team & same experimental setup
reproducible results
different team & different experimental setup
reproducible inferences
similar conclusions from independent replication or re-analysis
Git is version control
a way to preserve the history of changes
Edit & Save
one person or many people collaboratively
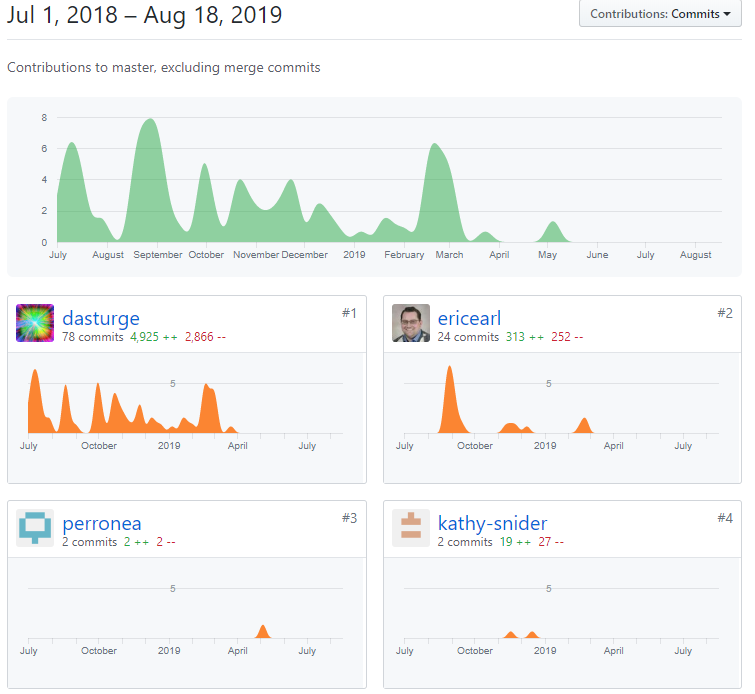
Reproducible Git
anyone can make exact copies at exact versions
Scenario 1: Merging changes
I have processing pipeline code that is missing a new option.
Kathy has already developed and tested that code out.
We can work together to merge Kathy's changes into my version.
Scenario 2: Restoring old stuff
Anders changed some analysis code to add a feature.
His new change breaks Oscar's old analysis.
Oscar can restore the old version and avoid the new changes.
DEMO: GitHub in action
- Use Search to find the ABCD-STUDY GitHub organization
- Open the abcd-dicom2bids repository
- Browse the src folder
- Go back to the top of the repo and Clone or download
JupyterLab is a lab notebook
a way to record and repeat exactly what you did before
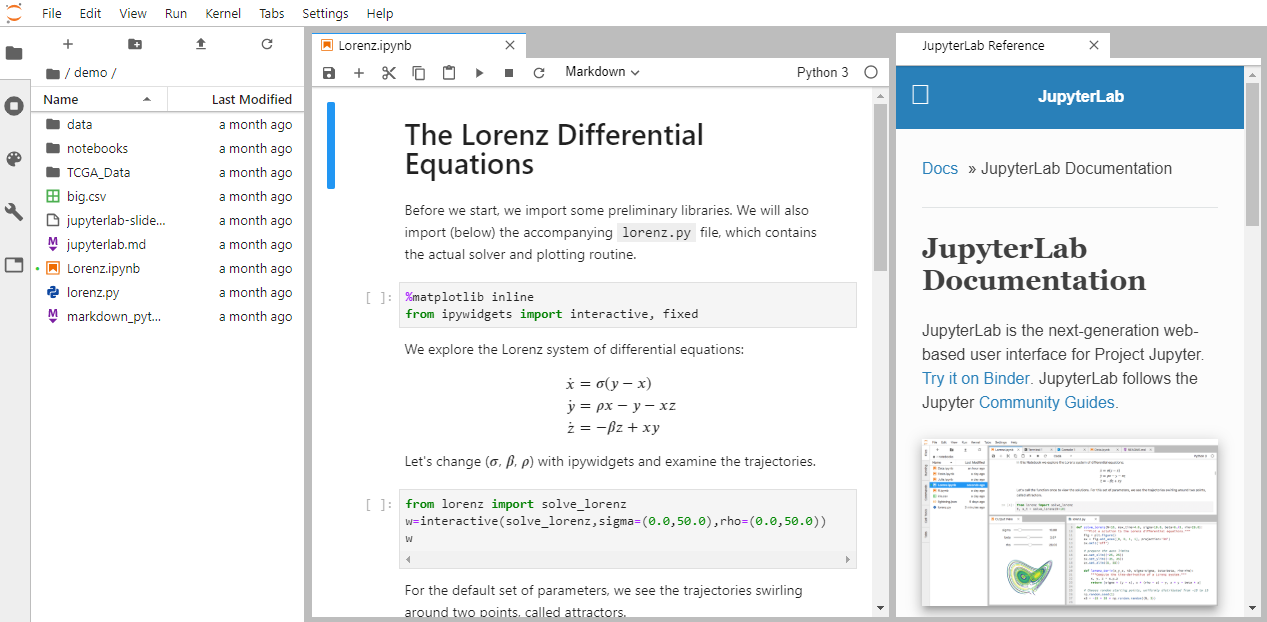
Text, code, and graphics
JupyterLab notebooks work interactively

DEMO: JupyterLab in action
- Open JupyterLab online
- Notice the difference between Markdown and Code cells
- Run the first two Code cells
Docker is containerization
containers (a.k.a. images) package dependencies and environments




Pull from DockerHub
containers built from Linux operating systems
docker pull dcanlabs/abcd-hcp-pipeline
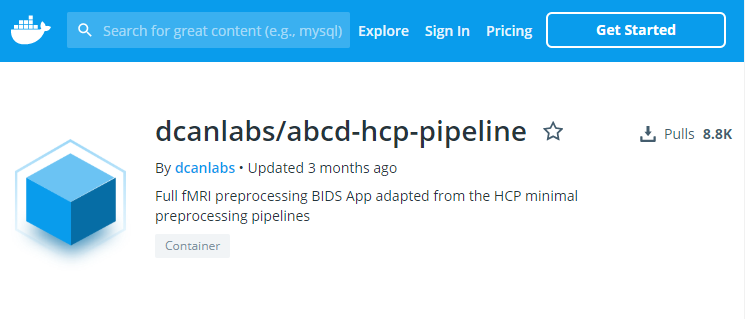
Exact environments
share your code without duplicating environments
Scenario 1: Collaboration
I have a processing pipeline and a server cluster.
My collaborator cross-country has a laptop.
No complicated setup and configuration, only need to run Docker.
Scenario 2: Reproducible results
Different team, different compute setup.
Mac, Linux, or Windows.
Container always behaves the same.
Running Containers
docker run [DOCKER_OPTIONS] IMAGE[:TAG] [CMD] [CMD_ARG(S)...]
### COMMON OPTIONS AND THEIR MEANINGS ###
# -it Get an interactive terminal #
# --rm Clean up container on exit #
# -v /A:/B Mount /A inside image as /B #
#########################################
# Open an Ubuntu 18.04 Docker image with a BASH terminal
# and have your home folder available inside as /myhome
docker run -it --rm \
-v ${HOME}:/myhome \
ubuntu:18.04 /bin/bashBIDS is a growing standard
the standard defines file/folder structure, data, and metadata
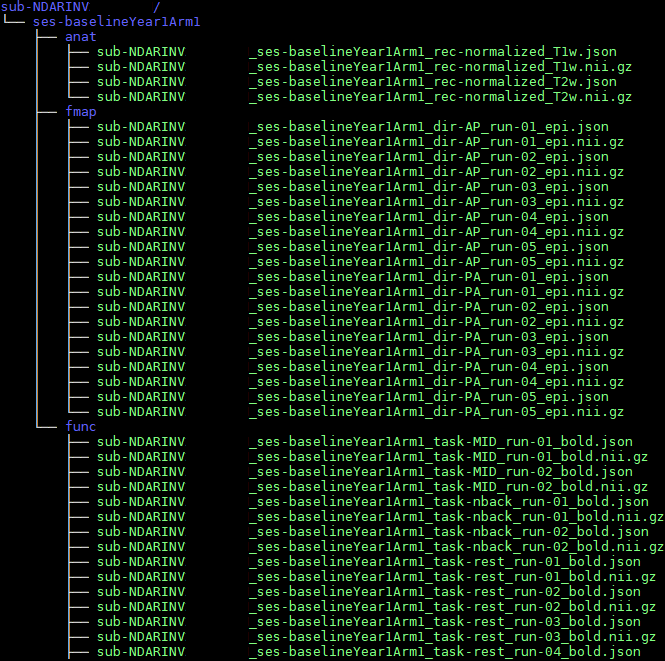
Data & Metadata
for every data file there is a metadata file
{
"Modality": "MR",
"MagneticFieldStrength": 3,
"Manufacturer": "Siemens",
"ManufacturersModelName": "Prisma",
"DeviceSerialNumber": "anon8928",
"BodyPartExamined": "BRAIN",
"PatientPosition": "HFS",
"SoftwareVersions": "syngo_MR_E11",
"MRAcquisitionType": "3D",
"SeriesDescription": "ABCD-T1-NORM_SIEMENS_original_(baseline_year_1_arm_1)",
"ProtocolName": "ABCD_T1w_MPR_vNav",
"ScanningSequence": "GR_IR",
"SequenceVariant": "SK_SP_MP",
"ScanOptions": "IR_WE",
"SequenceName": "tfl3d1_16ns",
"ImageType": [
"ORIGINAL",
"PRIMARY",
"M",
"ND",
"NORM"
],
"SeriesNumber": 5,
Different studies
have the same layout
no matter the study, I can still find imaging data
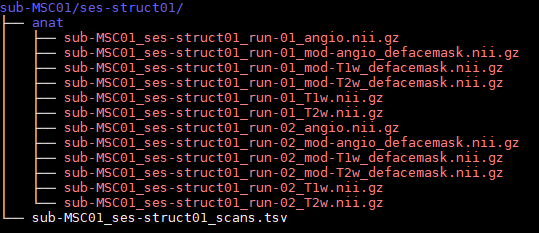
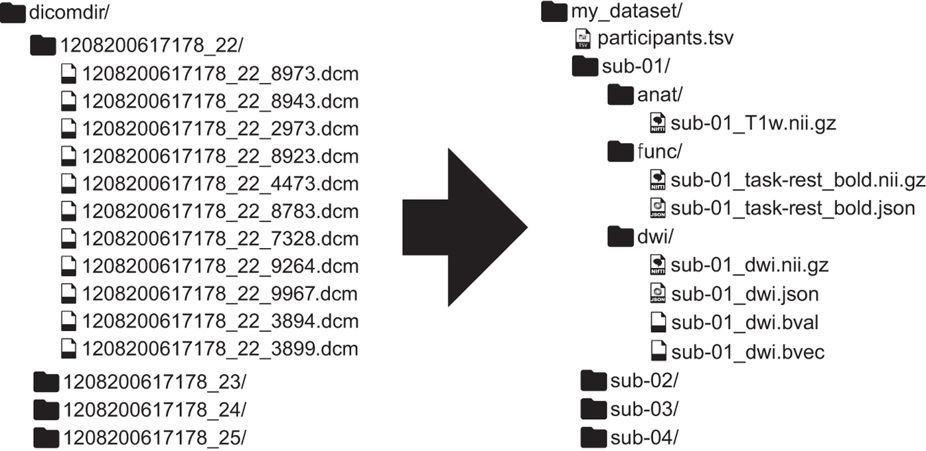
Dcm2Bids converts DICOMs
go from raw MRI data directly to BIDS
Only data you want
a config file allows you flexibility in what to convert
{
"descriptions": [
{
"dataType": "anat",
"modalityLabel": "T1w",
"criteria": {
"SeriesDescription": "ABCD-T1_SIEMENS_original_(baseline_year_1_arm_1)"
}
},
{
"dataType": "func",
"modalityLabel": "bold",
"customLabels": "task-rest",
"criteria": {
"SeriesDescription": "ABCD-rsfMRI_SIEMENS_mosaic_original_(baseline_year_1_arm_1)"
}
}
]
}Regular BIDS setup
get all the BIDS MRI inputs with less hassle
# FIRST STEPS DIRECTLY FROM DCM2BIDS REPOSITORY
# 1. cd <YOUR_FUTURE_BIDS_FOLDER>
# 2. dcm2bids_scaffold
# 3. dcm2bids_helper -d <FOLDER_WITH_DICOMS_OF_A_TYPICAL_SESSION>
# 4. Build your configuration file with the help of the content
# of tmp_dcm2bids/helper
# For the dcm2bids command itself:
# DICOM_DIR is a directory of DICOMs
# PARTICIPANT_ID and SESSION_ID are IDs picked by you
# These IDs MUST be only alphanumeric with no symbols
# CONFIG_FILE is a Dcm2Bids configuration JSON file
# Read here for more on CONFIG_FILE:
# https://cbedetti.github.io/Dcm2Bids/config/
dcm2bids -d DICOM_DIR -p PARTICIPANT_ID -s SESSION_ID -c CONFIG_FILEApps are standardized tools
usage: run.py [-h]
[--participant_label PARTICIPANT_LABEL [PARTICIPANT_LABEL ...]]
bids_dir output_dir {participant,group}
Example BIDS App entry point script.
positional arguments:
bids_dir The directory with the input dataset formatted
according to the BIDS standard.
output_dir The directory where the output files should be stored.
If you are running a group level analysis, this folder
should be prepopulated with the results of
the participant level analysis.
{participant,group} Level of the analysis that will be performed. Multiple
participant level analyses can be run independently
(in parallel).
optional arguments:
-h, --help show this help message and exit
--participant_label PARTICIPANT_LABEL [PARTICIPANT_LABEL ...]
The label(s) of the participant(s) that should be
analyzed. The label corresponds to
sub-<participant_label> from the BIDS spec (so it does
not include "sub-"). If this parameter is not provided
all subjects will be analyzed. Multiple participants
can be specified with a space separated list.Many Apps
the community is making more apps all the time
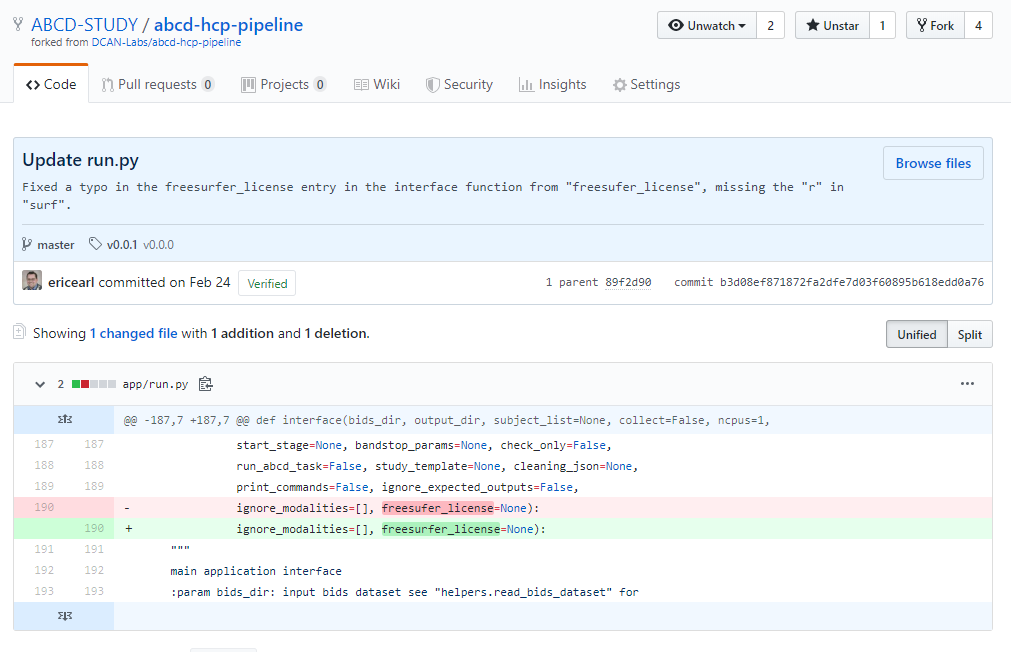
dcanlabs/abcd-hcp-pipeline
the pipeline used by the next big ABCD NDA data share
# Run the abcd-hcp-pipeline on all subjects
# within the local /path/to/bids_dataset
# mounted "read-only" (ro) as /input
# and /path/to/outputs as /output
# and /path/to/freesurfer/license
# as /license
docker run -it --rm \
-v /path/to/bids_dataset:/input:ro \
-v /path/to/outputs:/output \
-v /path/to/freesurfer/license:/license \
dcanlabs/abcd-hcp-pipeline /input /output \
--freesurfer-license=/license [OPTIONS]
standards are young and growing
standards for all output/derivative data types
ABCD-BIDS Derivatives
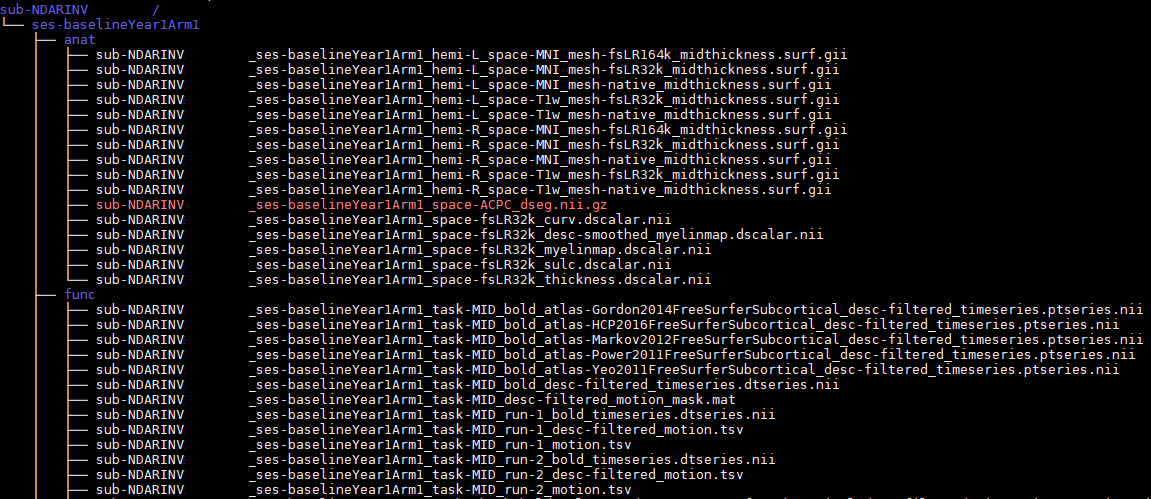
NDA is the ABCD data site
imaging and non-imaging data alike are hosted openly

DEMO: NDA in action
- Open NDA website
- Look at the Get Data in beta
- Look at Available Datasets and the (i) info button
ABCD-BIDS Collection
Collection #3165
DCAN Labs ABCD-BIDS MRI pipeline inputs and derivatives
estimated NDA release: September-November 2019
-
All ABCD Study participants' baseline imaging data that passed QC from the DAIC were processed by OHSU DCAN Labs
-
BIDS inputs and abcd-hcp-pipeline processed BIDS derivatives
Take-aways (revisited)
can you...
-
Find, browse, and clone GitHub repositories?
-
Open a JupyterLab Notebook and use it?
-
Run a Docker image (or a BIDS App)?
-
Navigate BIDS data and metadata?
-
Know where to find ABCD Study data?
My personal favorites
-
Good enough practices in scientific computing
Wilson, et al, PLOS Computation Biology, 2017.
-
The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments
Gorgolewski, et al, Scientific Data, 2016.
-
GitHub - "Built for developers", but realistically everybody...
-
BALSA File Types - Explanation of common imaging file formats
Thanks