data science
for (physical) scientists 8
dr.federica bianco | fbb.space | fedhere | fedhere


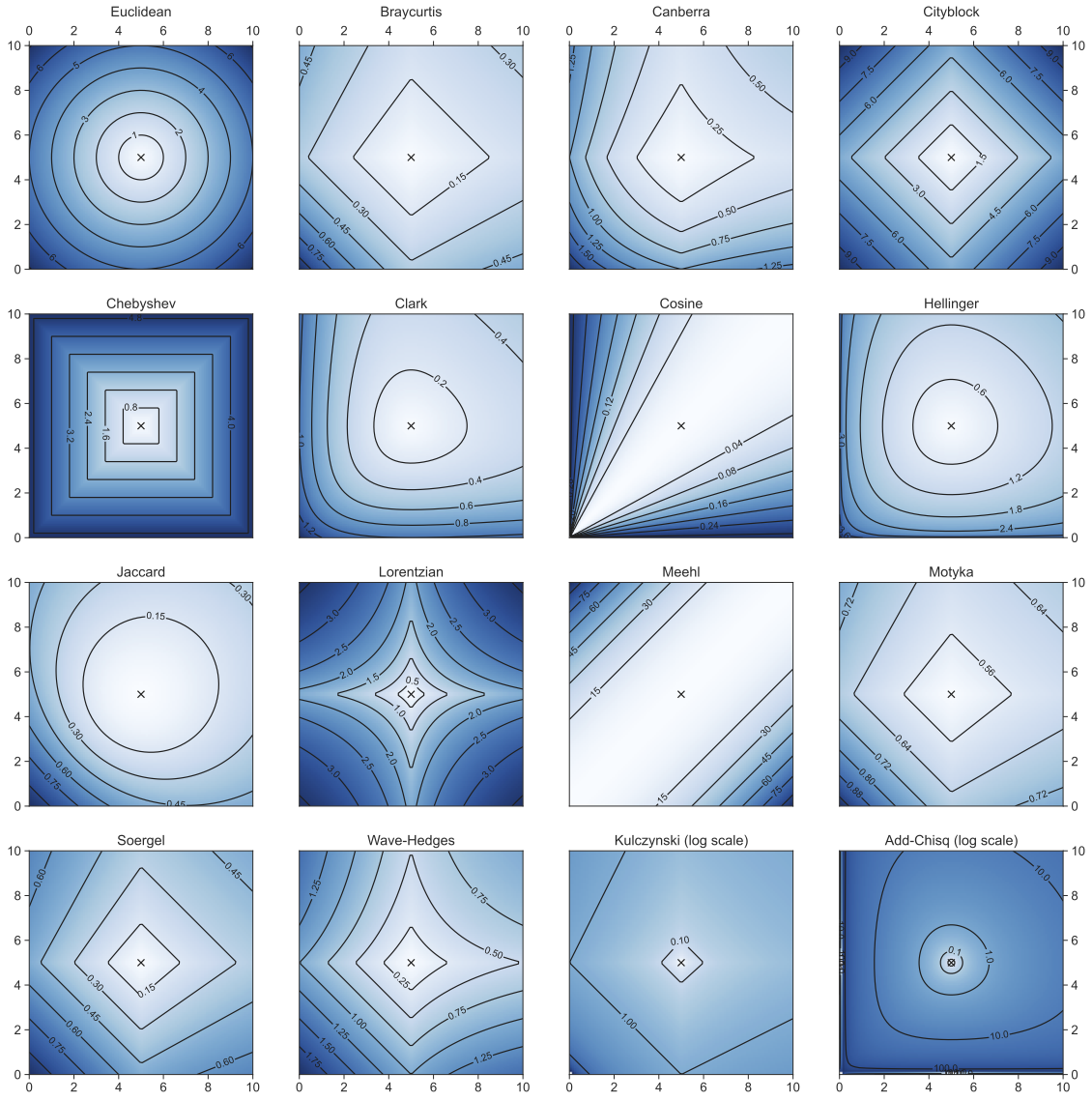
distances
this slide deck
k-Nearest Neighbor
0
Machine Learning
unsupervised learning
identify features and create models that allow to understand structure in the data
unsupervised learning
identify features and create models that allow to understand structure in the data
supervised learning
extract features and create models that allow prediction where the correct answer is known for a subset of the data
k-Nearest Neighbors
Calculate the distance d to all known objects Select the k closest objects Assign the most common among the k classes:
# k = 1
d = distance(x, trainingset)
C(x) = C(trainingset[argmin(d)])"lazy learner"
Calculate the distance d to all known objects Select the k closest objects
Classification:
Assign the most common among the k classes
Regression: Predict the average (median) of the k target values
k-Nearest Neighbors
Good
non parametric
very good with large training sets
Cover and Hart 1967: As n→∞, the 1-NN error is no more than twice the error of the Bayes Optimal classifier.
k-Nearest Neighbors
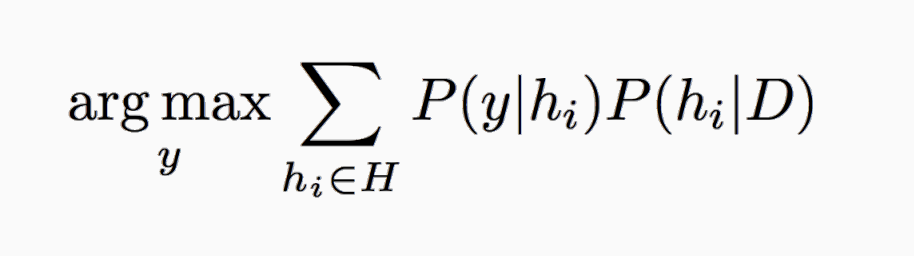
Good
non parametric
very good with large training sets
Cover and Hart 1967: As n→∞, the 1-NN error is no more than twice the error of the Bayes Optimal classifier.
Let xNN be the nearest neighbor of x.
For n→∞, xNN→x(t) => dist(xNN,x(t))→0
Theorem: e[C(x(t)) = C(xNN)]< e_BayesOpt
e_BayesOpt = argmaxy P(y|x)
Proof: assume P(y|xt) = P(y|xNN)
(always assumed in ML)
eNN = P(y|x(t)) (1−P(y|xNN)) + P(y|xNN) (1−P(y|x(t))) ≤
(1−P(y|xNN)) + (1−P(y|x(t))) =
2 (1−P(y|x(t)) = 2ϵBayesOpt,
k-Nearest Neighbors
Good
non parametric
very good with large training sets
Not so good
it is only as good as the distance metric
If the similarity in feature space reflect similarity in label then it is perfect!
poor if training sample is sparse
poor with outliers
k-Nearest Neighbors

k-Nearest Neighbors
Wine Example
Lazy Learning
PROS:
Because the model does not need to provide a global optimization the classification is "on-demand".
This is ideal for recommendation systems: think of Netflix and how it provides recommendations based on programs you have watched in the past.
CONS:
Need to store the entire training dataset (cannot model data to reduce dimensionality).
Training==evaluation => there is no possibility to frontload computational costs
Evaluation on demand, no global optimization - doesn’t learn a discriminative function from the training data but “memorizes” the training dataset instead.
distances
1
distance metrics
distance metrics
Any algorithm that fulfills the following conditions
distance metrics
continuous variables
Minkowski family of distances
distance metrics
continuous variables
Minkowski family of distances
N features (dimensions)
distance metrics
continuous variables
Minkowski family of distances
Manhattan: p=1
features: x, y
distance metrics
continuous variables
Minkowski family of distances

Manhattan: p=1
features: x, y
distance metrics
continuous variables
Minkowski family of distances

Euclidean: p=2
features: x, y
distance metrics
Minkowski family of distances
Great Circle distance
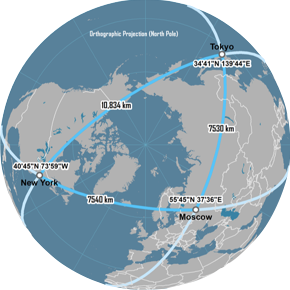
features
latitude and longitude
continuous variables
import scipy as sp
sp.spatial.distance.pdist(X) # the pairwise distance: returns (N**2 - N )/2 values for N objects
sp.spatial.distance.squareform(sp.spatial.distance.pdist(wines[["Alcohol", "Magnesium"]]))
#returns the NXN matrix of distances
plt.imshow(sp.spatial.distance.squareform(sp.spatial.distance.pdist(wines[["Alcohol", "Magnesium"]])))
#you can visualize the NXN matrix
plt.xlabel("wine")
plt.ylabel("wine");
plt.colorbar(label="distance");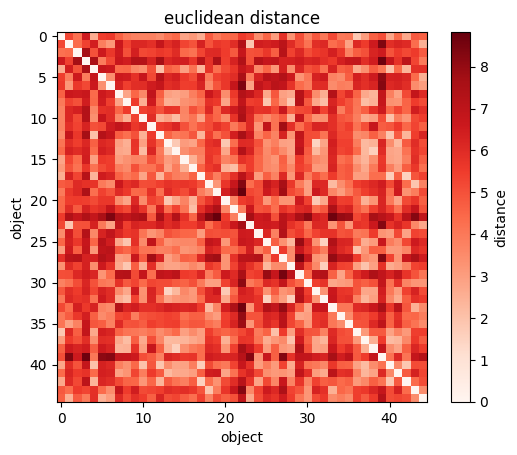
import scipy as sp
sp.spatial.distance.pdist(X) # the pairwise distance: returns (N**2 - N )/2 values for N objects
sp.spatial.distance.squareform(sp.spatial.distance.pdist(wines[["Alcohol", "Magnesium"]]))
#returns the NXN matrix of distances
plt.imshow(sp.spatial.distance.squareform(sp.spatial.distance.pdist(wines[["Alcohol", "Magnesium"]])))
#you can visualize the NXN matrix
plt.xlabel("wine")
plt.ylabel("wine");
plt.colorbar(label="distance");
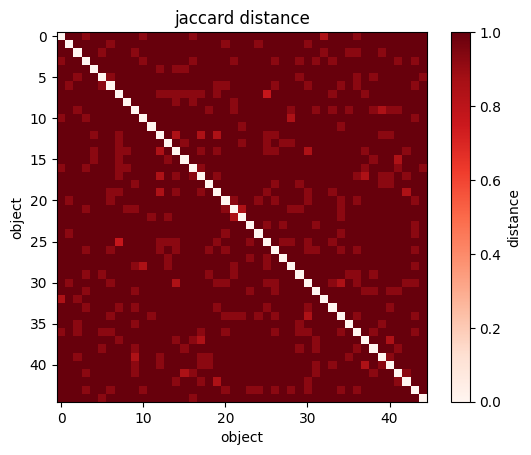
import scipy as sp
sp.spatial.distance.pdist(X) # the pairwise distance: returns (N**2 - N )/2 values for N objects
sp.spatial.distance.squareform(sp.spatial.distance.pdist(wines[["Alcohol", "Magnesium"]],
metric='jaccard'))
#returns the NXN matrix of distances
plt.imshow(sp.spatial.distance.squareform(sp.spatial.distance.pdist(wines[["Alcohol", "Magnesium"]])))
#you can visualize the NXN matrix
plt.xlabel("wine")
plt.ylabel("wine");
plt.colorbar(label="distance");
#Great Circle Distance in the sky
import astropy.units as u
from astropy.coordinates import SkyCoord
#The on-sky separation can be computed with the astropy.coordinates.BaseCoordinateFrame.separation()
#or astropy.coordinates.SkyCoord.separation() methods,
#which computes the great-circle distance (not the small-angle approximation):
c1 = SkyCoord('5h23m34.5s', '-69d45m22s', frame='icrs')
c2 = SkyCoord('0h52m44.8s', '-72d49m43s', frame='fk5')
sep = c1.separation(c2)Angle 20.74611448 deg
from shapely.geometry import Point
import geopandas as gpd
pnt1 = Point(80.99456, 7.86795)
pnt2 = Point(80.97454, 7.872174)
points_df = gpd.GeoDataFrame({'geometry': [pnt1, pnt2]}, crs='EPSG:4326')
points_df = points_df.to_crs('EPSG:5234')
points_df2 = points_df.shift() #We shift the dataframe by 1 to align pnt1 with pnt2
points_df.distance(points_df2)https://www.codedrome.com/calculating-great-circle-distances-in-python/
https://pypi.org/project/great-circle-calculator/
from math import radians, degrees, sin, cos, asin, acos, sqrt
def great_circle(lon1, lat1, lon2, lat2):
lon1, lat1, lon2, lat2 = map(radians, [lon1, lat1, lon2, lat2])
return 6371 * (acos(sin(lat1) * sin(lat2) + cos(lat1) *
cos(lat2) * cos(lon1 - lon2))) #kmdistance metrics
Uses presence/absence of features in data
: number of features in neither
: number of features in both
: number of features in i but not j
: number of features in j but not i
categorical variables:
binary
What is the distance between a leopard and a lizard?
- they both have tails
- only lizards have scales
- neither have wings


distance metrics
Uses presence/absence of features in data
: number of features in neither
: number of features in both
: number of features in i but not j
: number of features in j but not i
categorical variables:
binary
What is the distance between a leopard and a lizard?
- they both have tails
- only lizards have scales
- neither have wings
distance metrics
| 1 | 0 | sum | |
|---|---|---|---|
| 1 | M11 | M10 | M11+M10 |
| 0 | M01 | M00 | M01+M00 |
| sum | M11+M01 | M10+M00 | M11+M00+M01+ M10 |
observation i
}
}
| 0 | sum | ||
|---|---|---|---|
| 1 | M10 | M11+M10 | |
| 0 | M01 | M00 | M01+M00 |
| sum | M11+M01 | M10+M00 | M11+M00+M01+ M10 |
1
1
1
0
distance metrics
Simple Matching Distance
Uses presence/absence of features in data
: number of features in neither
: number of features in both
: number of features in i but not j
: number of features in j but not i
Simple Matching Coefficient
or Rand similarity
| 1 | 0 | sum | |
|---|---|---|---|
| 1 | M11 | M10 | M11+M10 |
| 0 | M01 | M00 | M01+M00 |
| sum | M11+M01 | M10+M00 | M11+M00+M01+ M10 |
observation i
observation j
}
}
categorical variables:
binary
| 0 | sum | ||
|---|---|---|---|
| 1 | M11 | M10 | M11+M10 |
| 0 | M01 | M00 | M01+M00 |
| sum | M11+M01 | M10+M00 | M11+M00+M01+ M10 |
1
1
1
0
lizard/leopard
distance metrics
Jaccard similarity
Jaccard distance
| 1 | 0 | sum | |
|---|---|---|---|
| 1 | M11 | M10 | M11+M10 |
| 0 | M01 | M00 | M01+M00 |
| sum | M11+M01 | M10+M00 | M11+M00+M01+ M10 |
observation i
observation j
}
}
categorical variables:
binary
lizard/leopard
distance metrics
Jaccard similarity
Jaccard distance
| 1 | 0 | sum | |
|---|---|---|---|
| 1 | M11 | M10 | M11+M10 |
| 0 | M01 | M00 | M01+M00 |
| sum | M11+M01 | M10+M00 | M11+M00+M01+ M10 |
observation i
observation j
}
}
categorical variables:
binary
distance metrics
Jaccard similarity
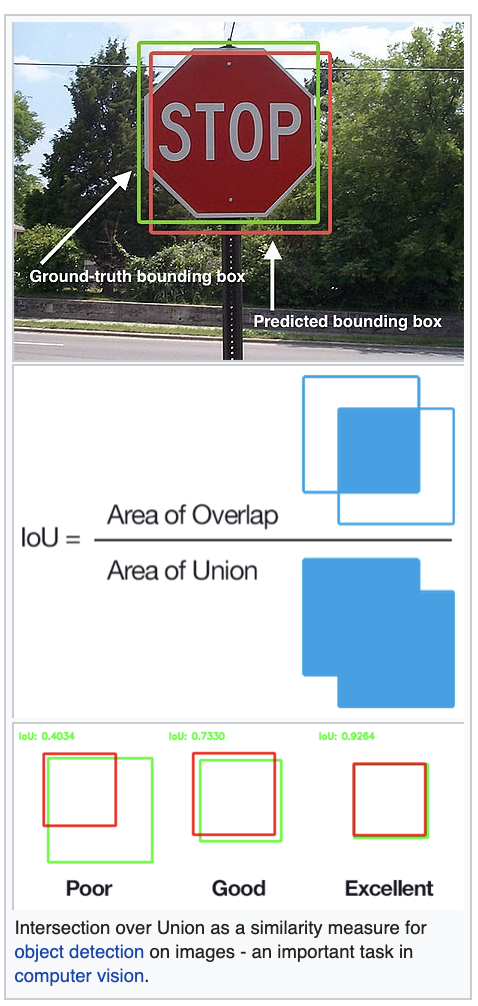
Application to Deep Learning for image recognition
Convolutional Neural Nets
categorical variables:
binary
distance metrics


3
whitening
Data can have covariance (and it almost always does!)
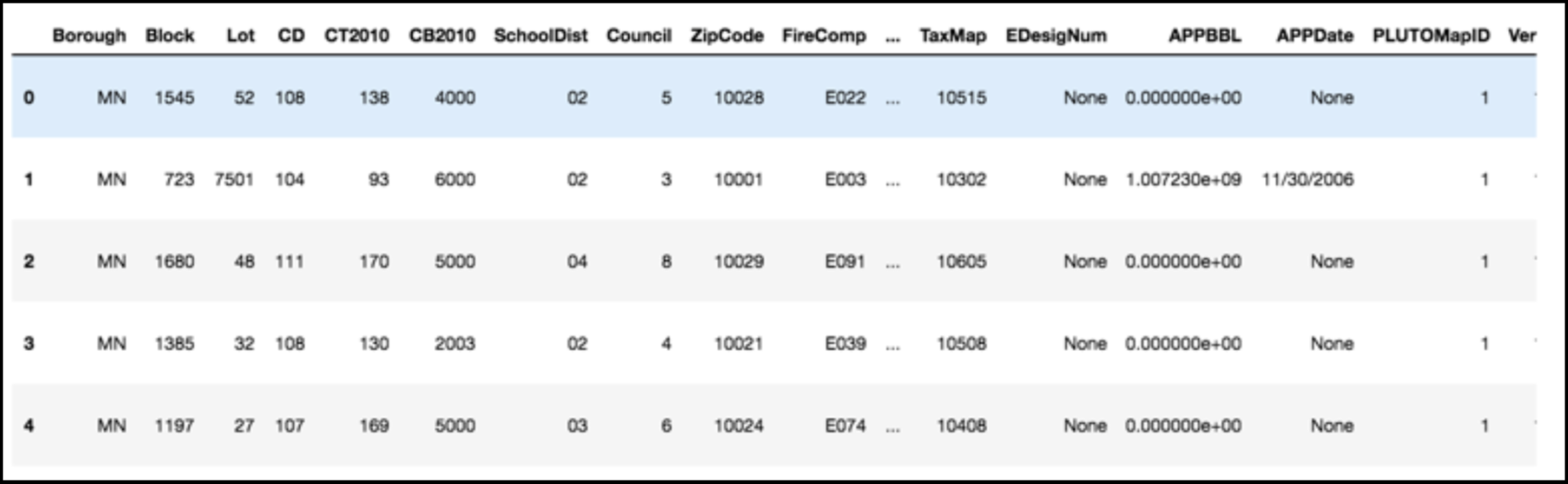
PLUTO Manhattan data (42,000 x 15)
axis 1 -> features
axis 0 -> observations
Data can have covariance (and it almost always does!)
PLUTO Manhattan data (42,000 x 15)
axis 1 -> features
axis 0 -> observations
COVARIANCE = correlation / variance
Data can have covariance (and it almost always does!)
Data can have covariance (and it almost always does!)
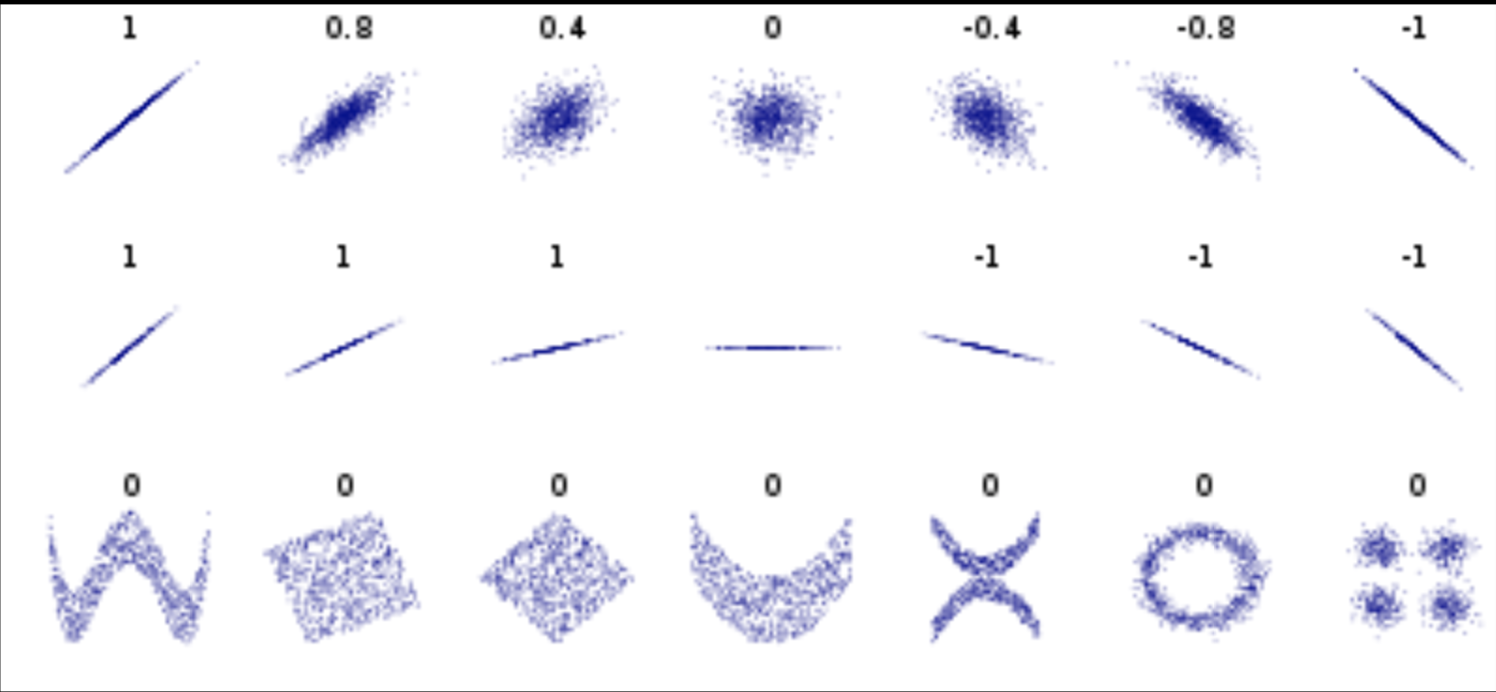
Pearson's correlation (linear correlation)
Generic preprocessing... WHY??
Worldbank Happyness Dataset https://github.com/fedhere/MLPNS_FBianco/blob/main/clustering/happiness_solution.ipynb
Clustering without scaling:
only the variable with more spread matters
Skewed data distribution:
std(x) ~ range(y)
Generic preprocessing... WHY??
Worldbank Happyness Dataset https://github.com/fedhere/MLPNS_FBianco/blob/main/clustering/happiness_solution.ipynb
Clustering without scaling:
only the variable with more spread matters


Skewed data distribution:
std(x) ~ range(y)

Clustering
Classifying &
regression
Unsupervised learning
- understanding structure
- anomaly detection
- dimensionality reduction
Supervised learning
- classification
- prediction
- feature selection
unsupervised vs supervised learning
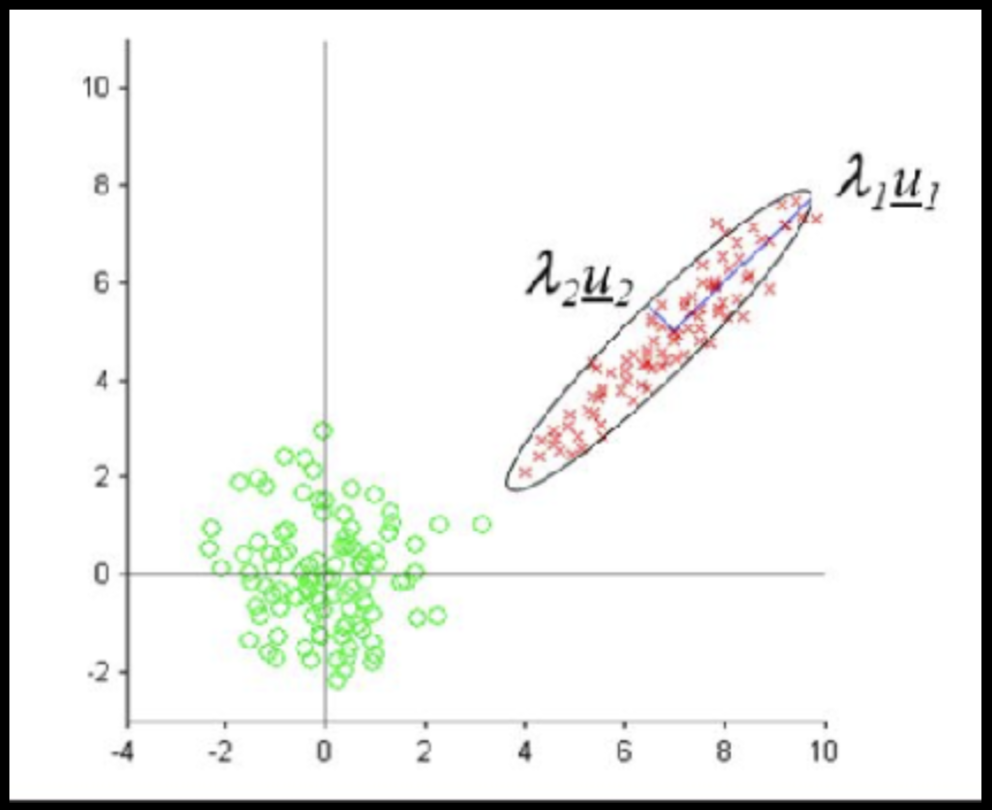
Data that is not correlated appear as a sphere in the Ndimensional feature space
Data can have covariance (and it almost always does!)
ORIGINAL DATA
STANDARDIZED DATA
Generic preprocessing
Generic preprocessing... WHY??
Worldbank Happyness Dataset
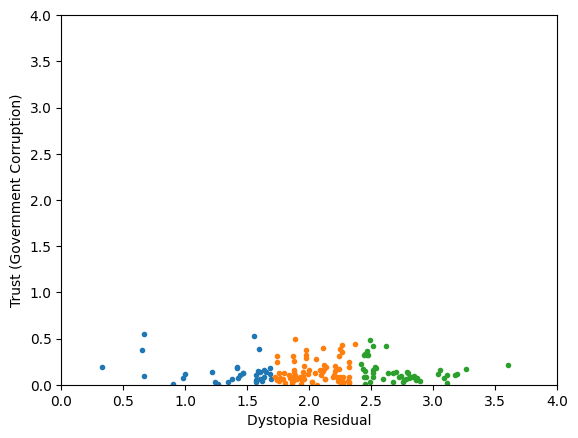
Classification/Clustering without scaling:
only the variable with more spread matters


Generic preprocessing... WHY??

Worldbank Happyness Dataset

Classification/Clustering without scaling:
only the variable with more spread matters
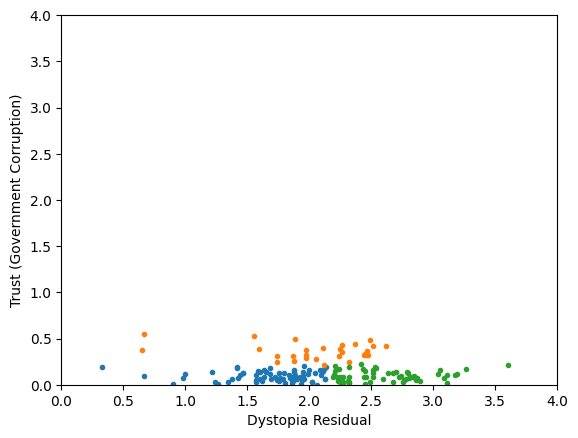
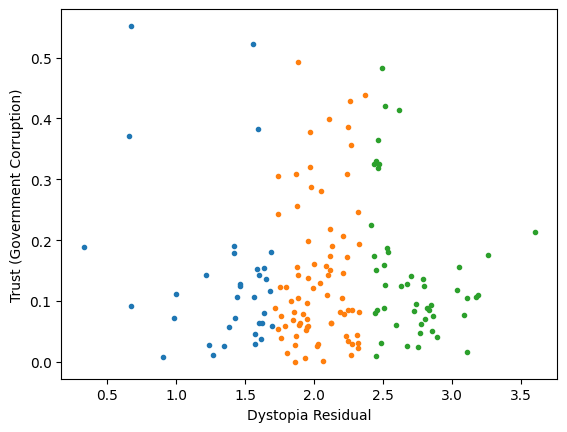
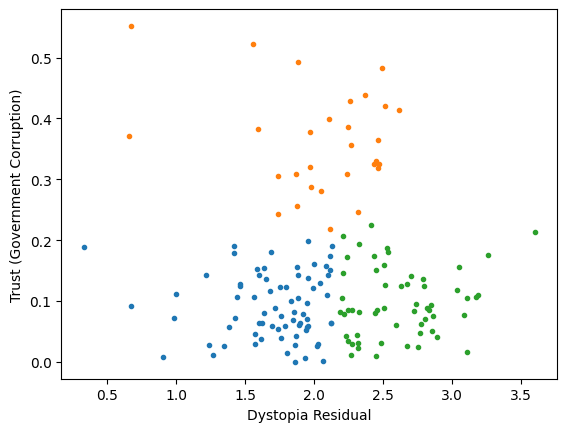
Classification/Clustering
after scaling:
both variables matter equally

Data that is not correlated appear as a sphere in the Ndimensional feature space
Data can have covariance (and it almost always does!)

ORIGINAL DATA
STANDARDIZED DATA
Generic preprocessing
Generic preprocessing
for each feature: divide by standard deviation and subtract mean
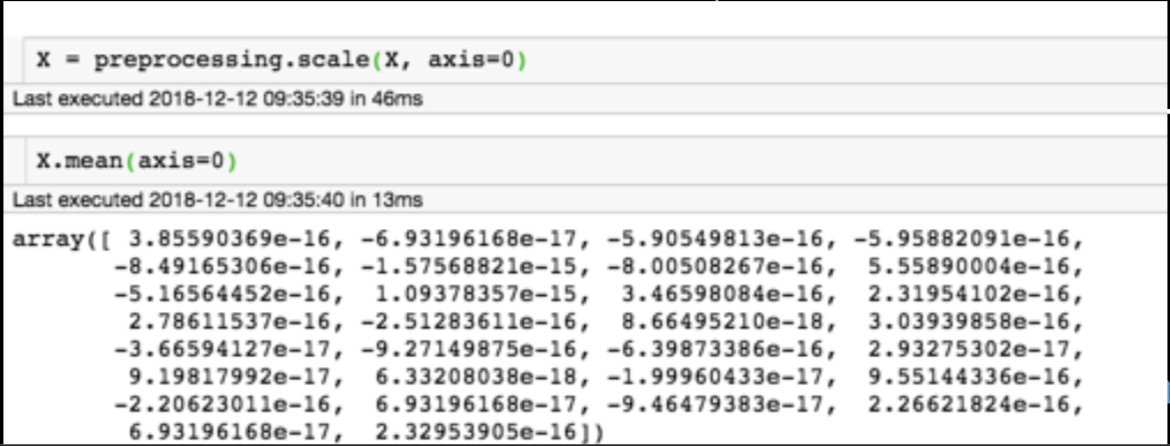
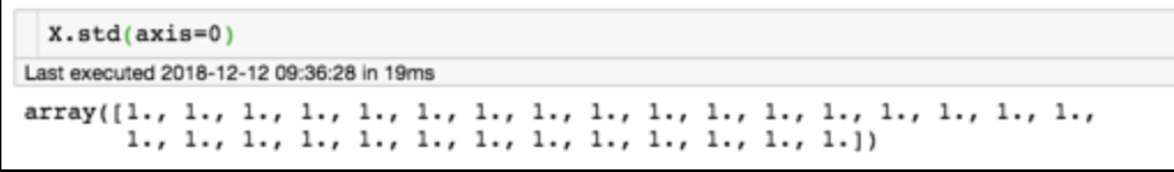

Generic preprocessing: most commonly, we will just correct for the spread and centroid
whitening
The term "whitening" refers to white noise, i.e. noise with the same power at all frequencies"
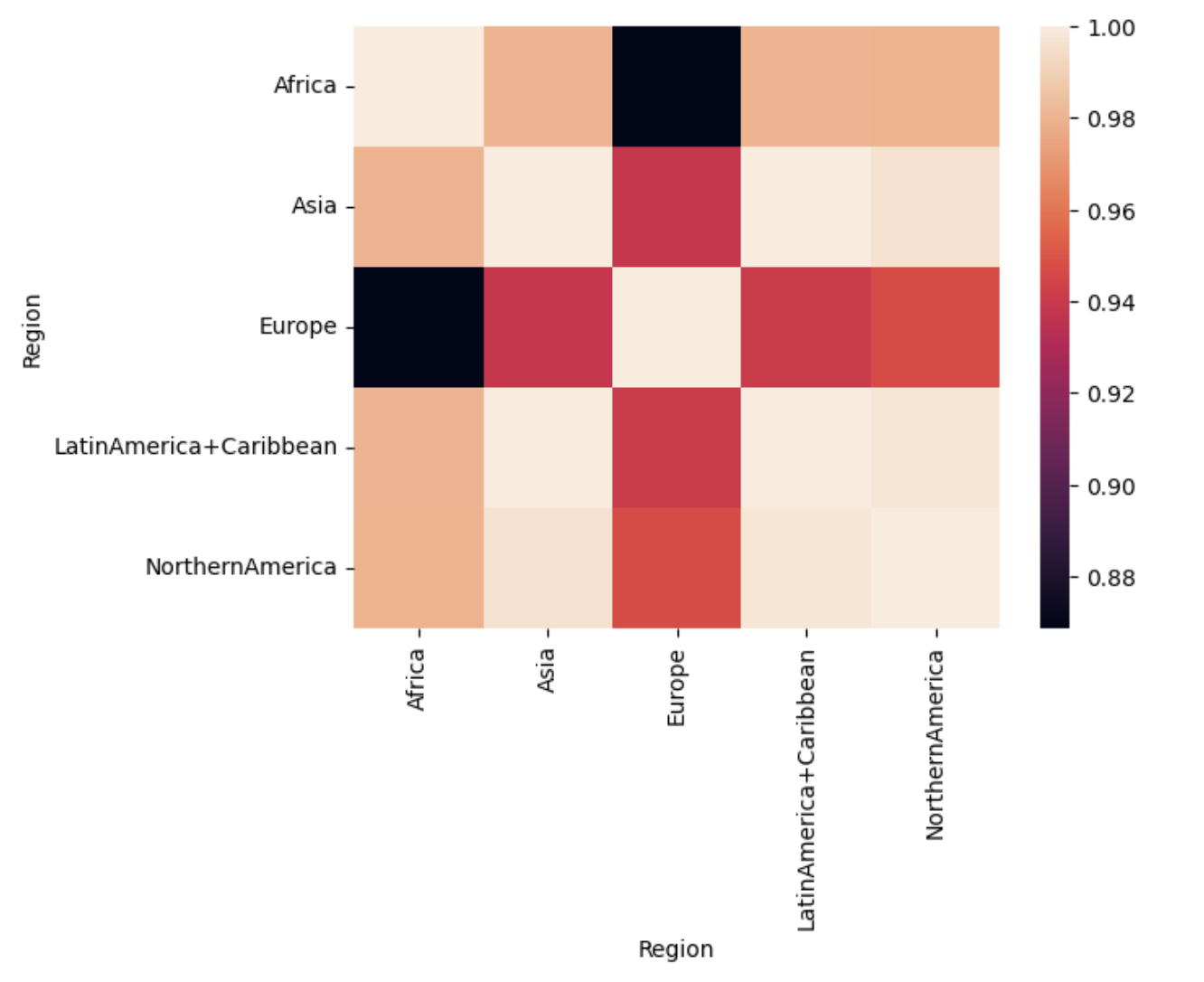
PLUTO Manhattan data (42,000 x 15) correlation matrix
axis 1 -> features
axis 0 -> observations
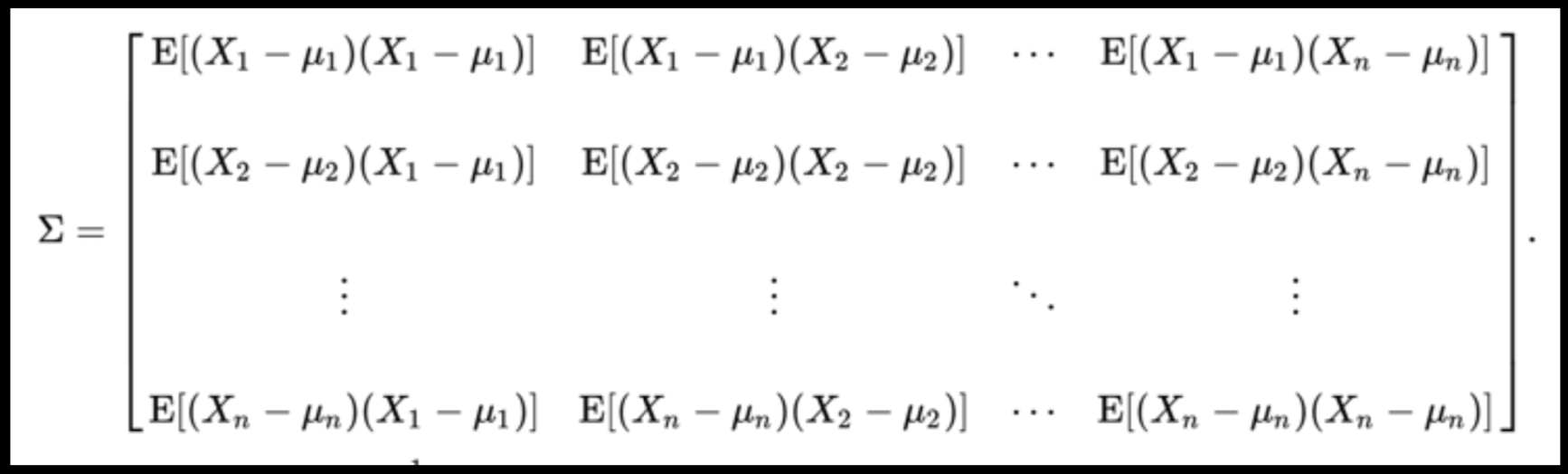
Data can have covariance (and it almost always does!)
PLUTO Manhattan data (42,000 x 15) correlation matrix
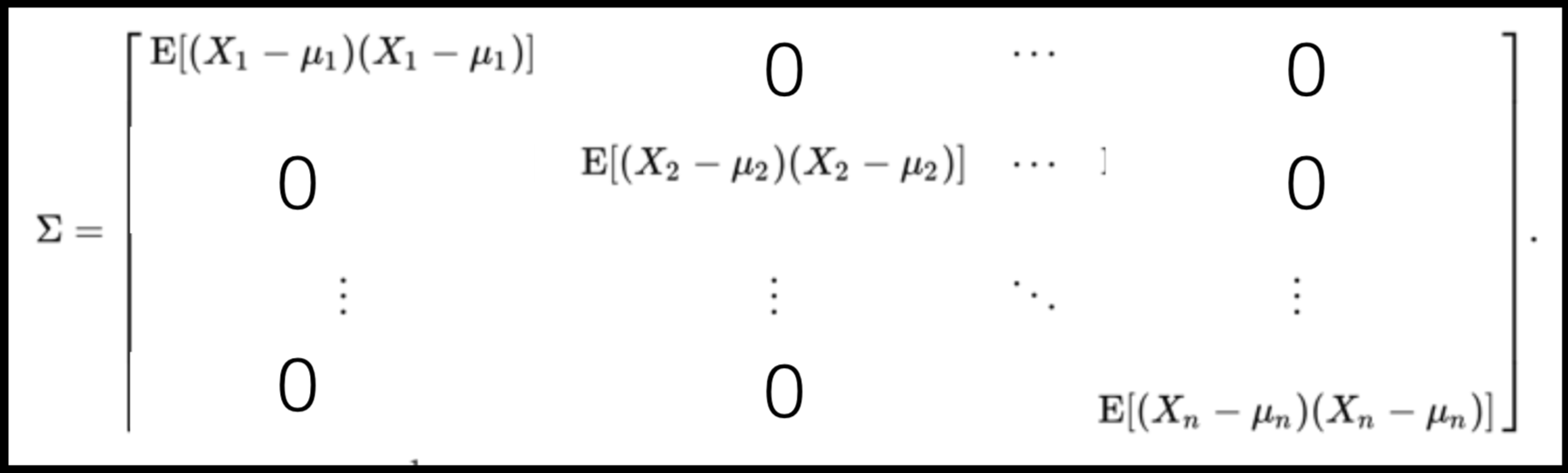
A covariance matrix is diagonal if the data has no correlation
Data can have covariance (and it almost always does!)
Full On Whitening
find the matrix W that diagonalized Σ
from zca import ZCA import numpy as np
X = np.random.random((10000, 15)) # data array
trf = ZCA().fit(X)
X_whitened = trf.transform(X)
X_reconstructed =
trf.inverse_transform(X_whitened)
assert(np.allclose(X, X_reconstructed))
: remove covariance by diagonalizing the transforming the data with a matrix that diagonalizes the covariance matrix
this is at best hard, in some cases impossible even numerically on large datasets
Generic preprocessing: other common schemes
for image processing (e.g. segmentation) often you need to mimmax preprocess
from sklearn import preprocessing
Xopscaled = preprocessing.minmax_scale(image_pixels.astype(float), axis=1)
Xopscaled.reshape(op.shape)[200, 700]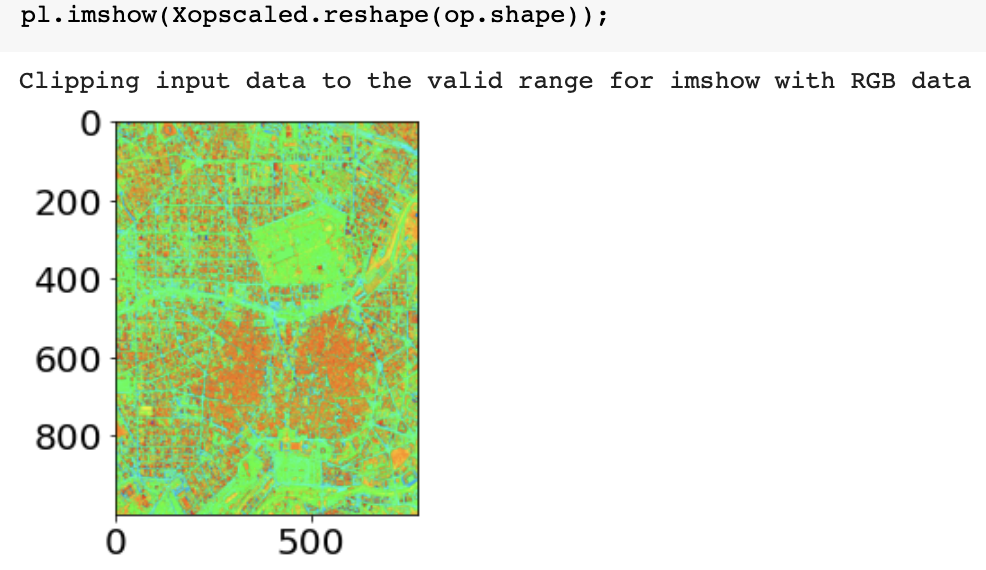

before
after (looks the same but colorbar different)
-107
273

0
1