Nonlinear mixed-effects models
Houssein Assaad, PhD
Senior Statistician and Software Developer
StataCorp LLC
SUGM, Colombus OH
19 Jul 2018

Outline
- What are nonlinear mixed-effects models ? Why and when you should use them ?
- Exploring the syntax: A decay model
- Covariance Structures: A Pharmacokinetics example
- Multilevel specification: A growth model
- Summary
- Nonlinear hierarchical models
- Goal: To describe a continuous response that evolves nonlinearly over time (or other condition) as a function of covariates, while taking into account the correlation among observations within the same subject.
- Nonlinear models are often based on a model for the mechanism producing the response. As a consequence, the model parameters in a nonlinear model generally have a natural physical interpretation.
- Nonlinear mixed-effects models (NLME), also known as:
- Nonlinear multilevel models
- Popular in agricultural, environmental, and biomedical applications. For example, NLME models have been used in drug absorption in the body, intensity of earthquakes, and growth of plants.
...First, an existential question:
Why would you want to use NLME models ?
Why nonlinear ?
Why mixed-effects ?
Let \(h_t\) be the expected height of a tree at time \(t\) which is modelled using a logistic growth model
- \(\beta_1\) is the asymptotic height as \(t\to\infty\)
Why nonlinear ?
- \(\beta_3\) is the time it takes for a tree to grow from 50% to about 73% of \( \beta_1\)
- \(\beta_2\) is the time at which the tree reaches half of its asymptotic height



- The logistic curve
is approximated very well
in the interval \([0.4, 1.6]\) by the \(5^{th}\) degree polynomial
- Polynomial approximation is unreliable outside the original interval.

- Suppose you have a dataset of tree heights measured over time
Why nonlinear ?
- Interpretability : Coefficients in polynomial model do not have any physical interpretation (e.g. what is the interpretation of the coefficient of \(t^4\) ?)
- Parsimony : Linear polynomial model uses twice as many parameters as the nonlinear model (logistic model)
- Validity beyond the observed range of the data
+---------------------+
| id X Y |
|---------------------|
| 1 14 5.6 |
| 1 21 9.6 |
| 1 42 15.5 |
|---------------------|
| 2 15 12.6 |
| 2 21 16.4 |
| 2 42 16.9 |
| 2 51 18.9 |
+---------------------+
- Data consists of \(j=1,\ldots,N\) subjects, each measured \(n_j\) times
Why mixed-effects ?
- \(\beta_1\), \(\beta_2\), and \(\beta_3\) may be subject-specific, e.g.
fixed effect
random effect
mixed effect
where
where
Why mixed effects ?
- Observations within the same subject are correlated
- Mixed-effects models allow us to account for the correlation of \(Y_{sj}\) and \(Y_{tj}\) by including in our model a random effect \(U_j\) specific to \(j^{th}\) subject
- The inclusion of random effects allow the parameters of interest (e.g. asymptotic growth ) to have subject-specific interpretations. Very important in pharmacokinetics, for example, where the goal is to derive dosage regimen for a specific patient.
2 observations on \(j^{th}\) subject
The Stata 15 menl command
Exploring the syntax
Fun example: Unicorn health study in Zootopia
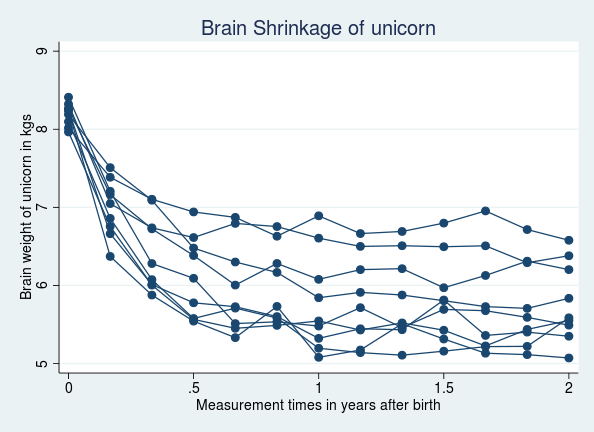
. webuse unicorn
(Brain shrinkage of unicorns in the land of Zootopia)
. twoway connected weight time if id < 10, connect(L) ///
title(Brain Shrinkage of unicorn)

+--------------------------+
| id time weight |
|--------------------------|
1. | 1 0 8.266746 |
2. | 1 .1666667 7.163768 |
3. | 1 .3333333 6.725025 |
4. | 1 .5 6.38325 |
5. | 1 .6666667 6.00481 |
|--------------------------|
776. | 60 1.333333 3.75486 |
777. | 60 1.5 3.581552 |
778. | 60 1.666667 3.701847 |
779. | 60 1.833333 3.836078 |
780. | 60 2 3.729806 |
+--------------------------+
. list id time weight if (_n <=5 | _n >775), sep(5)- Based on previous studies, a model for unicorn brain shrinkage is believed to be
where \(\epsilon_{ij}\sim N\left(0,\sigma^2\right)\)
- \(\beta_1\) is average asymptotic brain weight of unicorns as \(\texttt{time}_{ij} \to \infty\)
- \(\beta_2\) is average brain weight at birth (at \(\texttt{time}_{ij} = 0 \))
- \(\beta_3\) is a scale parameter that determines the rate at which the average brain weight approaches the asymptotic weight \(\beta_1\) (decay rate)
- We assume the asymptotic weight \(\beta_1\) is unicorn-specific (varies across unicorns) by including a random effect \(U_j\) as follows
- Thus our model may be written as
. menl weight = {b1}+{U[id]} + ({beta2} - ({b1}+{U[id]}) )*exp(-{beta3}*time)- Here is how you can fit it with menl
( Output is on next slide)
gsem-like syntax
. menl weight = {b1}+{U[id]} + ({beta2} - ({b1}+{U[id]}) )*exp(-{beta3}*time)
Obtaining starting values by EM:
Alternating PNLS/LME algorithm:
Iteration 1: linearization log likelihood = -56.9757597
Computing standard errors:
Mixed-effects ML nonlinear regression Number of obs = 780
Group variable: id Number of groups = 60
Obs per group:
min = 13
avg = 13.0
max = 13
Linearization log likelihood = -56.97576
------------------------------------------------------------------------------
weight | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
/b1 | 4.707954 .1414511 33.28 0.000 4.430715 4.985193
/beta2 | 8.089432 .0260845 310.12 0.000 8.038307 8.140556
/beta3 | 4.13201 .0697547 59.24 0.000 3.995293 4.268726
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
id: Identity |
var(U) | 1.189809 .2180058 .8308328 1.703887
-----------------------------+------------------------------------------------
var(Residual) | .0439199 .0023148 .0396095 .0486995
------------------------------------------------------------------------------
. menl weight = {b1}+{U[id]} + ({beta2} - ({b1}+{U[id]}) )*exp(-{beta3}*time)- We could have used the two-stage formulation of our model
. menl weight = {beta1:} + ({beta2} - {beta1:})*exp({beta3}*time), ///
define(beta1: {b1}+{U[id]})- Particularly useful for more complicated models
- Parameters specified within define() may be easily predicted after estimation
Incorporating covariates

- \(\texttt{cupcake}\) : number of rainbow cupcakes consumed right after birth
- \(\texttt{female}\) : \(\texttt{female}_j\) is \(1\) if the \(j\)th unicorn is a female and \(0\) otherwise
+---------------------------------------------+
| id cupcake time female weight |
|---------------------------------------------|
1. | 1 2 0 female 8.266746 |
2. | 1 2 .1666667 female 7.163768 |
3. | 1 2 .3333333 female 6.725025 |
4. | 1 2 .5 female 6.38325 |
5. | 1 2 .6666667 female 6.00481 |
|---------------------------------------------|
776. | 60 4 1.333333 male 3.75486 |
777. | 60 4 1.5 male 3.581552 |
778. | 60 4 1.666667 male 3.701847 |
779. | 60 4 1.833333 male 3.836078 |
780. | 60 4 2 male 3.729806 |
+---------------------------------------------+
. list if (_n <=5 | _n >775), sep(5)- Reducing brain weight loss has been an active research area in Zootopia, and scientists believe that consuming rainbow cupcakes right after birth may help slow down brain shrinkage.
- Also, we would like to investigate whether the asymptote, \(\beta_{1j}\), is gender specific, so we include the factor variable \(\mathtt{female}\) in the equation for \(\beta_{1j}\).
define(beta3: {b30} + {b31}*cupcake)define(beta1: {b10} + {b11}*1.female + {U[id]})decay rate
. menl weight = {beta1:} + ({beta2:} - {beta1:})*exp(-{beta3:}*time), ///
define(beta1: {b10}+{b11}*1.female+{U0[id]}) ///
define(beta2: {b2}) ///
define(beta3: {b30}+{b31}*cupcake)
-
Based on our results, consuming rainbow cupcakes after birth indeed slows down brain shrinkage: \(\mathtt{/b31}\) is roughly \(-0.2\) with a 95% \(\mathtt{CI}\) of \([-.27, -.13]\)
Mixed-effects ML nonlinear regression Number of obs = 780
Group variable: id Number of groups = 60
Obs per group:
min = 13
avg = 13.0
max = 13
Linearization log likelihood = -29.014988
beta1: {b10}+{b11}*1.female+{U0[id]}
beta2: {b2}
beta3: {b30}+{b31}*cupcake
------------------------------------------------------------------------------
weight | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
/b10 | 4.072752 .1627414 25.03 0.000 3.753785 4.39172
/b11 | 1.264407 .2299723 5.50 0.000 .8136694 1.715144
/b2 | 8.088102 .0255465 316.60 0.000 8.038032 8.138172
/b30 | 4.706926 .1325714 35.50 0.000 4.44709 4.966761
/b31 | -.2007309 .0356814 -5.63 0.000 -.2706651 -.1307966
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
id: Identity |
var(U0) | .7840578 .1438924 .5471839 1.123473
-----------------------------+------------------------------------------------
var(Residual) | .0420763 .0022176 .0379468 .0466551
------------------------------------------------------------------------------
Useful shortcut for your nonlinear expression
define(beta3: {b30} + {b31}*cupcake + {b32}*juice + {U2[id]})define(beta3: cupcake juice U2[id])- For more useful shortcuts, check the Random-effects substitutable expressions section in [ME] menl
define(beta3: {b30} + {b31}*cupcake)define(beta3: cupcake, xb)- What if you have many covariates ?
- Model parameters and random effects are bound in braces if specified directly in the expression: $$\texttt{\{b0\}}, \texttt{\{U0[id]\}} \text{, etc}.$$
- Linear combinations of variables can be included using the specification
- Random effects can be specified within a linear combination, in which case they should be included without curly braces, for example, $$\texttt{\{lc\_u: x1 x2 U[id]\}}$$
e.g.,
and
- Random-effects names must start with a capital letter as in
\(\texttt{\{U0[id]\}}\)
Pharmacokinetics (PK) study of Theophylline
- Pharmacokinetics is the study of “what the body does with a drug”

- PK seeks to understand how drugs enter the body, distribute throughout the body, and leave the body, by summarizing concentration-time measurements, while accounting for patient-specific characteristics.
- Maintain drug concentrations within the therapeutic window

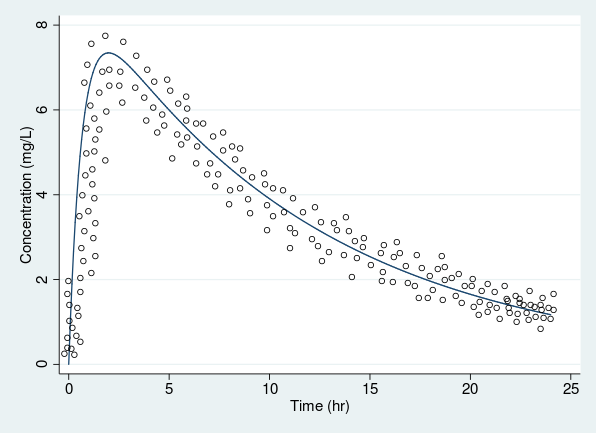
- Consider data on the antiasthmatic agent theophylline (Boeckmann, Sheiner, and Beal [1994] 2011).
- The drug was administrated orally to \(12\) subjects with the dosage (mg/kg) given on a per weight basis.
- Serum concentrations (in mg/L) were obtained at \(11\) time points per subject over \(25\) hours following administration.

. webuse theoph
. twoway connected conc time, connect(L)
- Clearance (\(\mathtt{Cl}\)) measures the rate at which a drug is cleared from the plasma. It is defined as the volume of plasma cleared of drug per unit time.
- Elimination rate constant (\(k_e\)) describes the rate at which a drug is removed from the body. It is defined as the fraction of drug in the body eliminated per unit time.
- Absorption rate constant (\(k_a\)) describes the rate at which a drug is absorbed by the body.
Because each of the model parameters must be positive to be meaningful, we write
(Output is on next slide)
. menl conc =(dose*{ke:}*{ka:}/({cl:}*({ka:}-{ke:})))* ///
(exp(-{ke:}*time)-exp(-{ka:}*time)), ///
define(cl: exp({b0}+{U0[subject]})) ///
define(ka: exp({b1}+{U1[subject]})) ///
define(ke: exp({b2}))
Mixed-effects ML nonlinear regression Number of obs = 132
Group variable: subject Number of groups = 12
Obs per group:
min = 11
avg = 11.0
max = 11
Linearization log likelihood = -177.02233
cl: exp({b0}+{U0[subject]})
ka: exp({b1}+{U1[subject]})
ke: exp({b2})
------------------------------------------------------------------------------
conc | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
/b0 | -3.227212 .0600089 -53.78 0.000 -3.344827 -3.109597
/b1 | .4656357 .1985941 2.34 0.019 .0763984 .854873
/b2 | -2.454679 .0524896 -46.77 0.000 -2.557556 -2.351801
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
subject: Independent |
var(U0) | .027864 .0121371 .0118652 .0654355
var(U1) | .4143467 .1945672 .165067 1.040082
-----------------------------+------------------------------------------------
var(Residual) | .5030242 .0688883 .3846079 .6578995
------------------------------------------------------------------------------
covariance(U1 U2 U3, unstructured)- By default, menl assumes that all random effects are independent, thus, in the previous model, we have assumed that \(\text{cov}\left(U_{0}, U_{1}\right) = 0\), but we could have considered other covariance structures for the random effects
- For example, when there are 3 random effects \(U_1\), \(U_2\), and \(U_3\) in the model, their covariance structure may be summarized as
covariance(U*, exchangeable)covariance(U*, identity)covariance(U2 U3, unstructured)- Other examples of random effects covariance structures are:
. menl conc =(dose*{ke:}*{ka:}/({cl:}*({ka:}-{ke:})))* ///
(exp(-{ke:}*time)-exp(-{ka:}*time)), ///
define(cl: exp({b0}+{U0[subject]})) ///
define(ka: exp({b1}+{U1[subject]})) ///
define(ke: exp({b2})) covariance(U*, unstructured)
- Going back to our PK model, we may wish to assume that \(U_{0j}\) and \(U_{1j}\) have a general covariance matrix
. menl conc =(dose*{ke:}*{ka:}/({cl:}*({ka:}-{ke:})))* ///
(exp(-{ke:}*time)-exp(-{ka:}*time)), ///
define(cl: exp({b0}+{U0[subject]})) ///
define(ka: exp({b1}+{U1[subject]})) ///
define(ke: exp({b2})) ///
covariance(U*, unstructured)
Linearization log likelihood = -177.02222
cl: exp({b0}+{U0[subject]})
ka: exp({b1}+{U1[subject]})
ke: exp({b2})
--------------------------------------------------------------------------------
conc | Coef. Std. Err. z P>|z| [95% Conf. Interval]
---------------+----------------------------------------------------------------
/b0 | -3.227224 .0600072 -53.78 0.000 -3.344836 -3.109612
/b1 | .4656428 .1985911 2.34 0.019 .0764113 .8548742
/b2 | -2.454696 .0524895 -46.77 0.000 -2.557574 -2.351819
--------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
subject: Unstructured |
var(U0) | .0278616 .0121451 .0118566 .0654714
var(U1) | .4143347 .194565 .1650595 1.040069
cov(U0,U1) | -.0002056 .0332183 -.0653122 .0649011
-----------------------------+------------------------------------------------
var(Residual) | .5030238 .0688885 .3846073 .6578995
------------------------------------------------------------------------------
- Combine resvariance() and rescorrelation() to build flexible covariance structures.
- Use option resvariance() to model heteroskedasticity as a linear, power, or exponential function of other covariates or of predicted values.
- Use option rescorrelation() to model the dependence of the within-group errors as, e.g., AR or MA processes.
- menl provides flexible modeling of within-group error or residual covariance structures.
So far, we have assumed that \(\text{Var}\left(\epsilon_{ij}\right) = \sigma^2\) and \(\text{cov}\left(\epsilon_{sj}, \epsilon_{tj}\right)=0\)
- Use option rescorrelation() to model the dependence of the within-group errors as, e.g., auto-regressive (AR) or moving-average (MA) processes.
Variance increases with mean

Constant variance
resvariance(power _yhat)- For example, heteroskedasticity, often present in PK data, is modeled using the power function plus a constant.
Adding a constant avoids the variance of zero at \(\texttt{time}_{ij}\) = 0, because the concentration is zero at that time.
- We will also use restricted maximum likelihood estimation (REML) via option reml instead of the default maximum likelihood (ML) estimation to account for a moderate number of subjects.
. menl conc =(dose*{ke:}*{ka:}/({cl:}*({ka:}-{ke:})))* ///
(exp(-{ke:}*time)-exp(-{ka:}*time)), ///
define(cl: exp({b0}+{U0[subject]})) ///
define(ka: exp({b1}+{U1[subject]})) ///
define(ke: exp({b2})) ///
resvariance(power _yhat) reml
Mixed-effects REML nonlinear regression
Linear. log restricted-likelihood = -172.44384
cl: exp({b0}+{U0[subject]})
ka: exp({b1}+{U1[subject]})
ke: exp({b2})
------------------------------------------------------------------------------
conc | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
/b0 | -3.227295 .0619113 -52.13 0.000 -3.348639 -3.105951
/b1 | .4354519 .2072387 2.10 0.036 .0292716 .8416322
/b2 | -2.453743 .0517991 -47.37 0.000 -2.555267 -2.352218
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
subject: Independent |
var(U0) | .0316416 .014531 .0128634 .0778326
var(U1) | .4500585 .2228206 .1705476 1.187661
-----------------------------+------------------------------------------------
Residual variance: |
Power _yhat |
sigma2 | .1015759 .086535 .0191263 .5394491
delta | .3106636 .2466547 -.1727707 .7940979
_cons | .7150935 .3745256 .2561837 1.996063
------------------------------------------------------------------------------
. predict (Cl = {cl:})
. list subject Cl if subject[_n]!=subject[_n-1], sep(0) noobs
- We can predict the clearance parameter \(\mathtt{Cl}_j\) for each subject
- Interpretation: For subject 1, \(0.027\) liters of serum concentration are cleared of the theophylline drug per hour per kg body weight.
- In other words, if subject 1 weighs 75 kg, \(75\times0.027 \approx 2\) liters of serum concentration are cleared of theophylline every hour
+--------------------+
| subject Cl |
|--------------------|
| 1 .0274995 |
| 2 .0402322 |
| 3 .0406919 |
| 4 .0371865 |
| 5 .0425807 |
| 6 .0461769 |
| 7 .0466375 |
| 8 .0442402 |
| 9 .0324682 |
| 10 .0356245 |
| 11 .0510018 |
| 12 .03785 |
+--------------------+
Summary II
Stage I:
Stage II:
menl y = <menlexpr>, define() [define() ...]covariance(U0 U1 [U2 ...], <covtype>)resvariance() rescorrelation()with
and
2-stage specification
Single-stage specification
menl y = <menlexpr>rescovariance()What about multilevel NLME models ?
- Consider fictional data on tree circumference of 30 trees planted in 5 different planting zones. The tree circumferences were recorded over the period of 1990 to 2003.
Zone 1
Zone 5
Tree 27
Tree 6
Tree 30
Tree 1
Tree 2
\(1990\)
\(1991\)
\(2003\)
Zone 2
Level 3
Level 2
Level 1
- We wish to fit the 3-parameter logistic growth model:
- \(\texttt{circum}_{ijt}\) and \(\texttt{age}_{ijt}\) are circumference (cm) and age (in years) of tree \(j\) within planting zone \(i\) in year \(t\)
- Parameters: Asymptotic circumference (\(\beta_1\)), time at which half of \(\beta_1\) is attained (\(\beta_2\)), and scale parameter (\(\beta_3\))
- For simplicity we assume that only \(\beta_2\) may be affected by zones and trees-within-zones:
. menl circum = {b1}/(1 + exp(-(age - {beta2:})/{b3})), ///
define(beta2: {b2} + {U1[zone]} + {U2[zone > tree]}) stddeviations
Mixed-effects ML nonlinear regression Number of obs = 420
-------------------------------------------------------------
| No. of Observations per Group
Path | Groups Minimum Average Maximum
----------------+--------------------------------------------
zone | 5 56 84.0 112
zone>tree | 30 14 14.0 14
-------------------------------------------------------------
Linearization log likelihood = 313.18701
beta2: {b2}+{U1[zone]}+{U2[zone>tree]}
------------------------------------------------------------------------------
circumf | Coef. Std. Err. z P>|z| [95% Conf. Interval]
-------------+----------------------------------------------------------------
/b1 | 30.01228 .0228861 1311.38 0.000 29.96742 30.05713
/b2 | 6.459742 .0757833 85.24 0.000 6.311209 6.608274
/b3 | 2.902458 .0046722 621.21 0.000 2.893301 2.911615
------------------------------------------------------------------------------
------------------------------------------------------------------------------
Random-effects Parameters | Estimate Std. Err. [95% Conf. Interval]
-----------------------------+------------------------------------------------
zone: Identity |
sd(U1) | .1599475 .057271 .0792858 .3226707
-----------------------------+------------------------------------------------
zone>tree: Identity |
sd(U2) | .1244983 .0178957 .0939312 .1650126
-----------------------------+------------------------------------------------
sd(Residual) | .0966751 .0034615 .0901233 .1037034
------------------------------------------------------------------------------- The time to reach half of the asymptotic circumference (\(\beta_2\)) for \(j\)th tree within planting zone \(i\):
. predict (beta2_i = {beta2:}), relevel(zone)
. list zone beta2_i if zone[_n]!=zone[_n-1], sep(0) noobs
+----------------------------+
| zone beta2_i |
|----------------------------|
| New England 6.357335 |
| Mid Atlantic 6.273229 |
| E North Central 6.723731 |
| W North Central 6.481954 |
| South Atlantic 6.46246 |
+----------------------------+
- We can predict the time to reach half of the asymptotic circumference (\(\beta_2\)) for each planting zone \(i\):
Summary
- menl fits NLME models; see [ME] menl
- menl supports ML and REML estimation and provides flexible random-effects and residual covariance structures.
-
menl supports single-stage and two-stage specifications.
- You can predict random effects and their standard errors, group-specific nonlinear parameters, and more after estimation; see [ME] menl postestimation.
- NLME models are known to be sensitive to initial values. menl provides default, but for some models you may need to specify your own. Use option initial().
- NLME models are known to have difficulty converging or converging to a local maximum. Trying different initial values may help.
- It also supports specifying models with no random effects (nonlinear marginal models)
ALWAYS BE YOURSELF
UNLESS YOU CAN BE A UNICORN
THEN ALWAYS BE A UNICORN

