Towards Accreditation in Metagenomics for Clinical Microbiology
Thesis Comittee
Programa de Doutoramento do Centro Académico de Medicina de Lisboa
Inês Mendes
15th of September, 2020
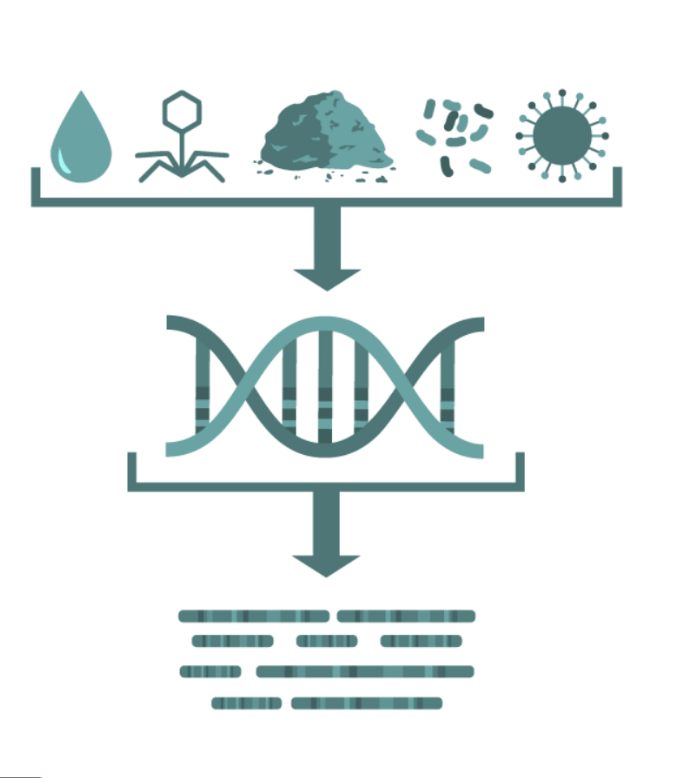
Metagenomics
Random "shotgun" sequencing of microbial DNA, without selecting a particular gene.
Promising methodology for obtaining fast results for the identification of pathogens and their virulence and antimicrobial resistance properties without the need for culture.
| The motivation
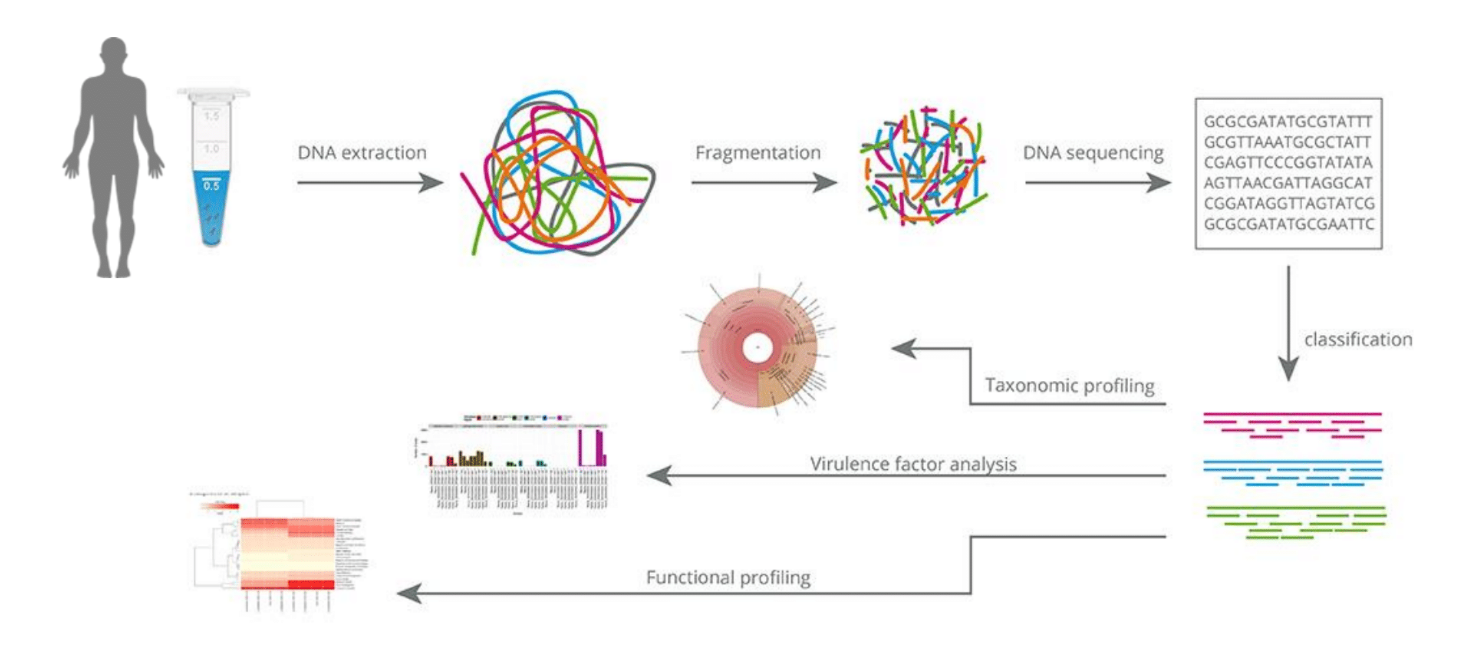
Who is there? - Taxonomic identification
What are they doing? - Virulome, Resistome, Functional Annotation
Who is doing what? - Functional Assignment
Metagenomics
| The motivation
- Development of novel methods and metrics to accurately identify and estimate relative abundance of viral and baterial pathogens.
- Standardizing the process of metagenomic analysis.
- Development of a computational efficient and robust framework.
Metagenomics
| The motivation
Main Goals
- Fundação para a Ciência e a Tecnologia (SFRH/BD/129483/2017)
- Dr. João A. Carriço - Instituto de Medicina Molecular, MRamirez Lab
- Prof. Dr. John W. A. Rossen - University Medical Center Groninen, Department of Medical Microbiology
Metagenomics
| The motivation
Funding
Promoters & Host Institutions

| Analysis
Metagenomics

Precision, Sensibility & Performance
(Culture + Maldi-TOF)
11 Metagenomic Samples - Fluid & Tissue
| Analysis
Metagenomics
- Highlights the potential and the limitations of shotgun metagenomics as a diagnostic tool - Lack of reproducibility
- Results are highly dependent on the tools, and specially database, chosen for the analysis - Lack of standardization and propper benchmark.
| Analysis
Metagenomics
Major issues
Reproducibility in Computational Biology
Clinical Shotgun Metagenomic Analysis
Programa de Doutoramento do Centro Académico de Medicina de Lisboa
Inês Mendes
15th of September, 2020
Reproducibility
| The motivation


- Do I know what is happening?
- Is it reproducible?
- Is it shareable?

Black
Box
Transparent
Box
- Commercial/Freeware
- You get what it gives you
- Ready to use
- Stealth change
- Standalone
- Freeware
- You can "tailor"
- "Major" headache
- Visible change
- Dependencies
The needs:
- Analyze a large amount of sequence data routinely
- Some computationally intensive steps
- Constantly updating/adding software
Reproducibility
| The motivation
Writing of pipelines in python/perl/shell scripts circa 2000, colorized.

- Custom ad-hoc scripts
- Difficult to parallelise
- Difficult to install/run
- Hard to deploy in multiple environments
- What's workflow managers?!
- What's docker?!
Workflows in the Paleolithic era:
Reproducibility
| The motivation
The game changing combination of workflow managers and containers:
- Portability
- Reproducible
- Scalability
- Multi-scale containerization
- Native cloud support







Reproducibility
| The motivation
Workflows in the Modern era:




Monolithic pipelines


Need to change often
Siloed tool containers
Don't do much by themselves
Reproducibility
| The motivation

Reproducibility
|




Workflow based development
Component based development
Components are modular pieces with some basic rules:

Component A
- Input/Output
- Parameters
- Resources
Component B

- Input/Output
- Parameters
- Resources
Reproducibility
|
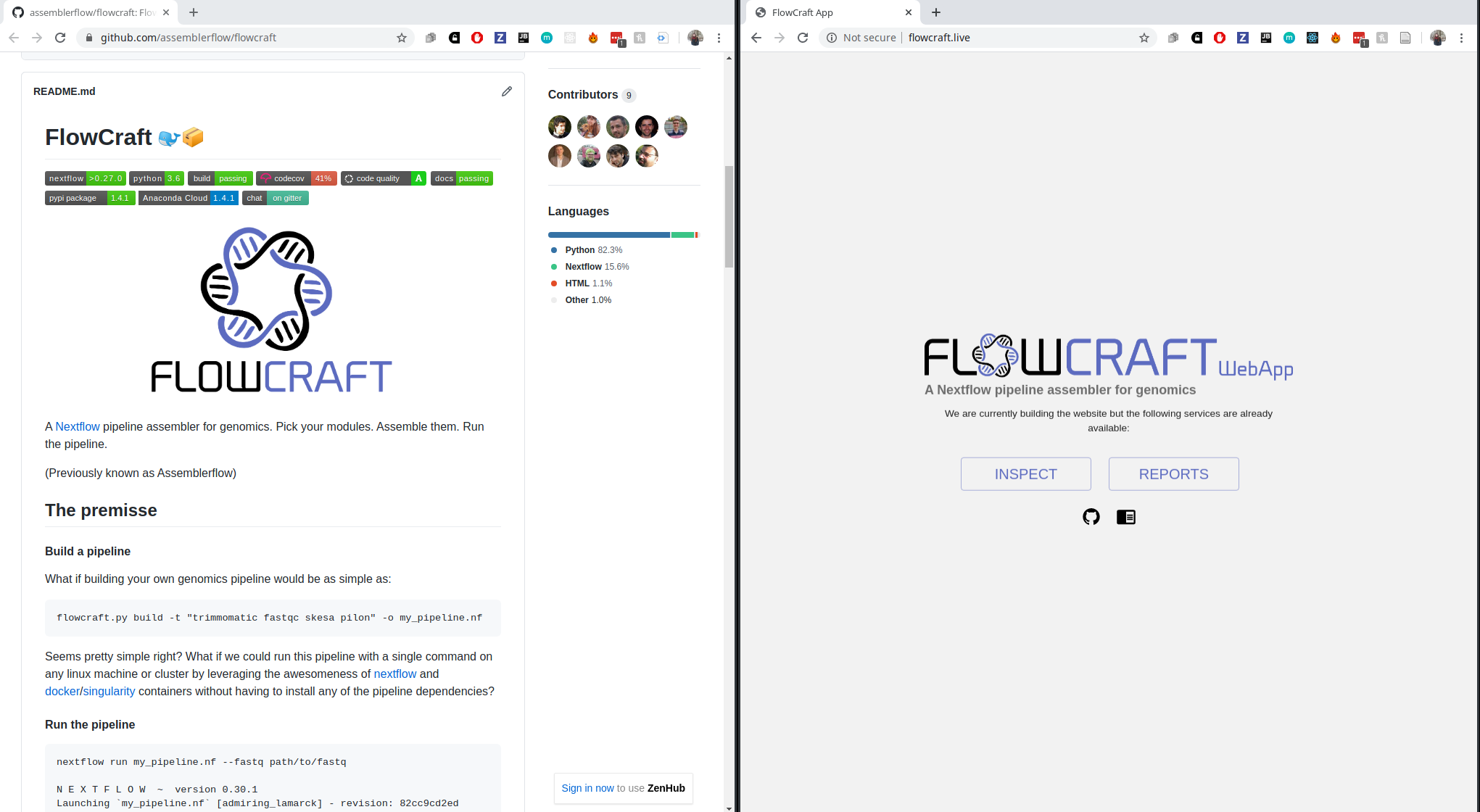
With this framework, building workflows becomes simple:
flowcraft build -t 'trimmomatic fastqc spades pilon' -o my_nextflow_pipeline
Results in the following workflow DAG (direct acyclic graph)
It's easy to get experimental:
flowcraft build -t 'trimmomatic fastqc skesa pilon' -o my_nextflow_pipeline
Switch spades for skesa

Reproducibility
|
Forks
Connect one component to multiple
Secondary channels
Connect non-adjacent components
Extra inputs
Inject user input data anywhere
Recipes
Curated and pre-assembled pipelines for specific needs

Reproducibility
|

Reproducibility
|

Reproducibility
|
Multiple Raw Input Types
Not limited to paired-end FastQ or Fasta
Dynamic Input in Components
One component, multiple inputs
Expand Building Features
New merge operators
- Component that receives multiple input types
- Component that merges multiple inputs from different branches

Reproducibility
|

Programa de Doutoramento do Centro Académico de Medicina de Lisboa
Inês Mendes
15th of September, 2020
dengue virus genotyping from amplicon and shotgun metagenomic sequencing

| The motivation


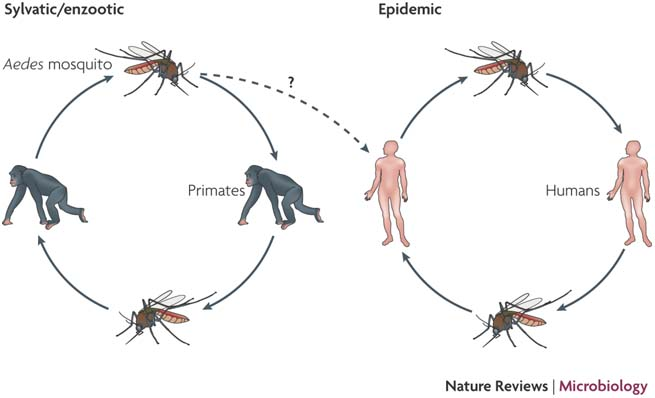
doi:10.1038/nrmicro1690
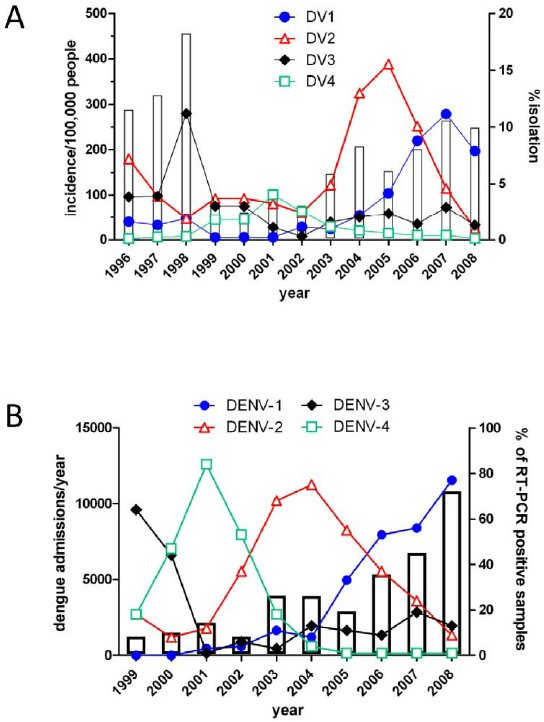
Sequential infection increases the risk of a severe form of the infection - dengue hemorrhagic fever.

| The motivation

Dengue hemorrhagic fever:
- The virus presents 4 different serotypes, sub-classified in many genotypes
- After "primary" infection with one serotype, "secondary" infections by one or more of the other serotypes can precipitate ‘antibody dependant enhancement’ (ADE)
- Infection with one type gives little immune protection against the other types
| The motivation

- DENV-1 - genotypes I-V
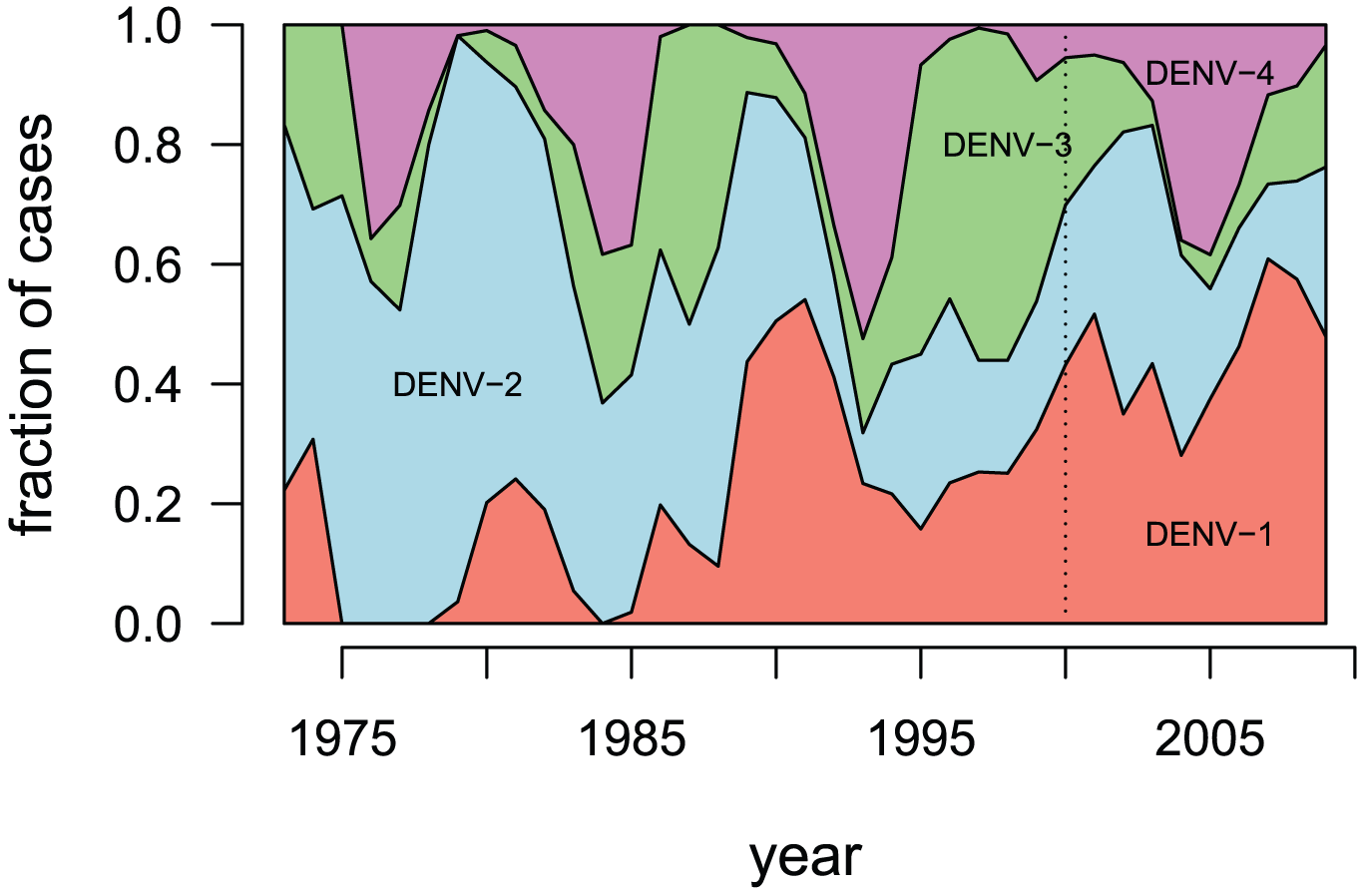
- DENV-2 - genotypes Asian I, Asian II, Cosmopolitan, American, Asian/American & Sylvatic
- DENV-3 - genotypes I-V
- DENV-4 - genotypes I - III & Sylvatic
https://doi.org/10.1371/journal.pntd.0001876.g002
https://doi.org/10.1371/journal.pntd.0000757
Thailand
Viet Nam
| The motivation

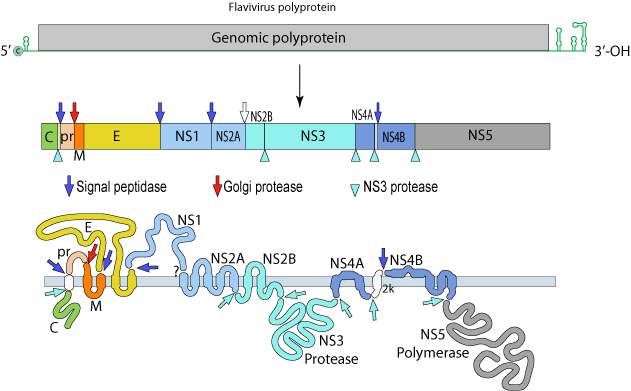
DENV: (+)ssRNA (~11Kb; 1 ORF)
The single polyprotein encodes:
-
Structural Proteins:
-
C – capsid
-
prM – pre-membrane
-
M - membrane
-
E - envelope
-
-
Non-Structural Proteins:
-
NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5
-

| The motivation

Empower the use of HTS to monitor the dissemination of the disease
RNA Extraction
PCR
Amplification
HTS Sequencing
➔
➔

How do the different genotypes model transmission and infection?
| The motivation


| The workflow

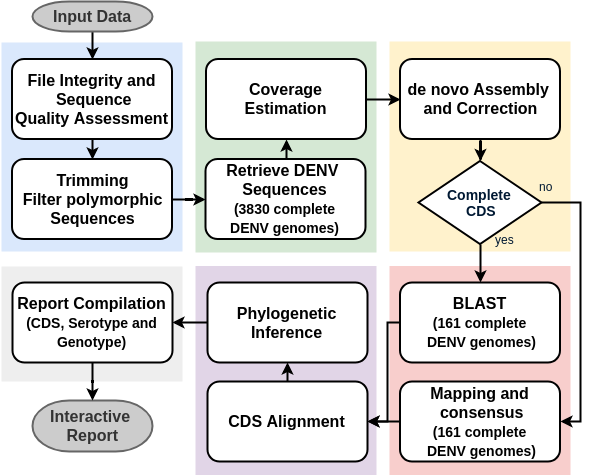




- Ready to use but customizable
- Scalable
- Reproducible
- Stand-alone (confidential data)
- Easy to explore and share results
Requirements
A solution
| The workflow



DENV Identification
- 3830 complete DENV genomes from the NIAID Virus Pathogen Database and Analysis Resource (ViPR)
- complete genome sequence
- human & mosquito host (exception of DENV-1 III, monkey)
- collection year (1950-2019)
In Silico Typing:
- 161 representative sequences of all sero and genotypes.
| The workflow


- DENV-1


- DENV-2
a) Envelope Region b) whole genome sequence
| DENV Classification

a) Envelope Region b) whole genome sequence
- DENV-3


- DENV-4
| DENV Classification

Shotgun Metagenomics dataset:
- 9 plasma samples
- 13 serum samples
- 1 spiked sample with the 4 serotypes
- Positive and Negative controls


nextflow run DEN-IM.nf -profile slurm_shifter --fastq="fastq/*_{1,2}.*"- HPC with 300 Cores/Processing Power and 3 TB RAM
- On average, each sample took 7 minutes to analyse. A total of 75 CPU hours were used to analyse the 25 samples, with a total of 17Gb in size. This analysis resulted in 69Gb of data.

| Use case

| Use case




Git, Nextflow (java) and a container engine (Docker, singularity, shifter...).
apt-get install gitcurl -s https://get.nextflow.io | bash
apt-install docker-ce
Clone (or run remotely)
git clone https://github.com/B-UMMI/DEN-IM.githttps://github.com/B-UMMI/DEN-IM/wiki| Installation
(Meta)Genomic Assembly Benchmark
de novo Assembly of short-read data
Programa de Doutoramento do Centro Académico de Medicina de Lisboa
Inês Mendes
15th of September, 2020
The assembly methods provide longer sequences that are more informative than shorter sequencing data and can provide a more complete picture of the microbial community in a given sample.
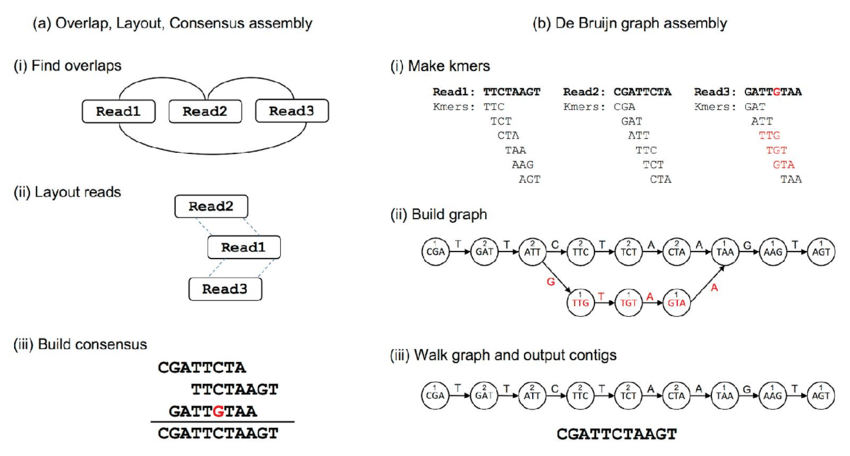
Benchmark
| (Meta)Genomic assembly
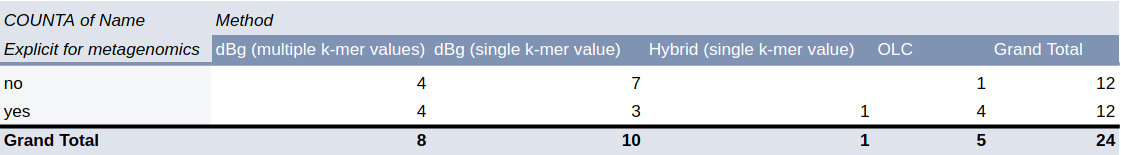
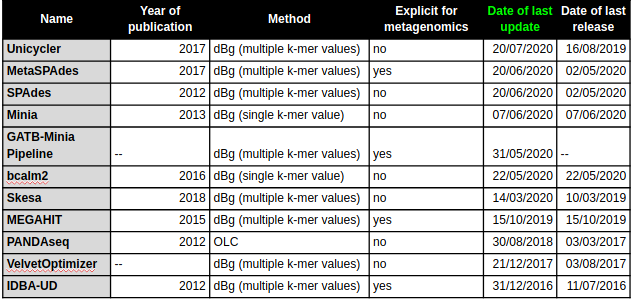
Benchmark
| (Meta)Genomic assembly
Benchmark
| (Meta)Genomic assembly
Benchmark
| Assembly workflow
Reference Dataset (Complete Bacterial Genomes)
In silico mock sample (even)
In silico mock sample (log)
Zymos standard (even)
Zymos standard (log)
3.7 M read pairs
8.8 M read pairs
47.8 M read pairs
Assembly Workflow
Assembly Quality Assessment



Benchmark
| Assembly evaluation
Reference Dataset (Triple)

Assembly file (fasta)
Filter min contig size (1000 bp)
Mapping with Minimpa2
Read Data

PAF file (tab)
Benchmark
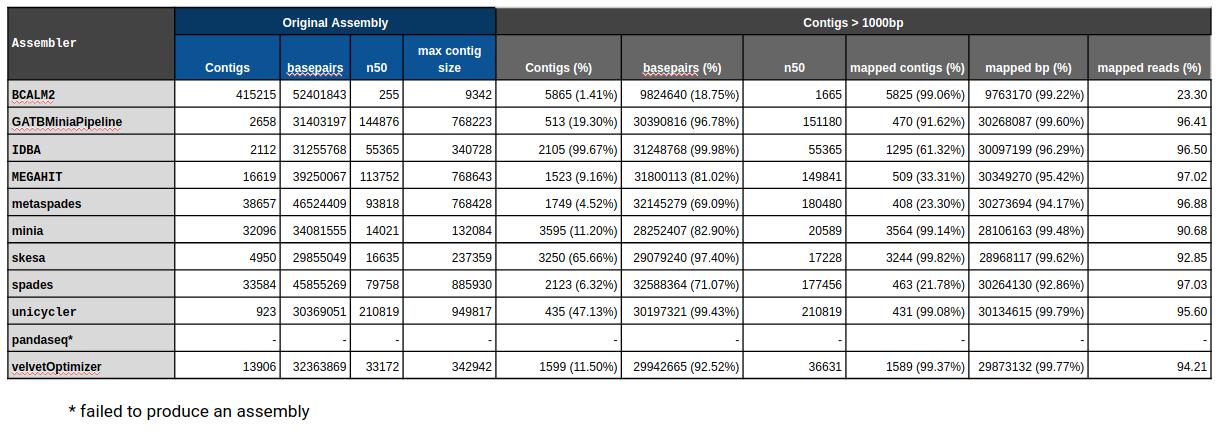
| Assembly evaluation
General Assembly & Global Mapping Statistics
Mapping Statistics & Metrics per Reference
- Number of contigs
- Number of basepairs
- N50
- Max contig size
- Number of contigs (%)
- Number of basepairs (%)
- N50
- % Mapped contigs
- % Mapped basepairs
- % Mapped reads
Original
Filtered (1000bp)
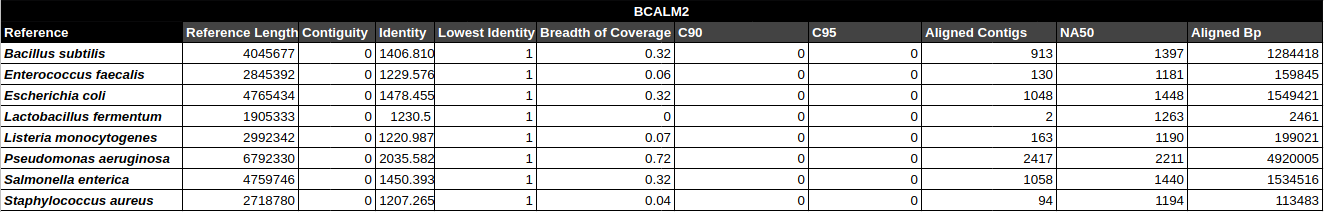
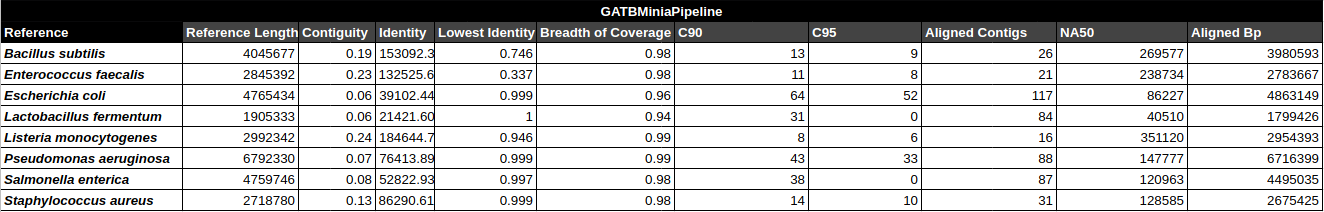
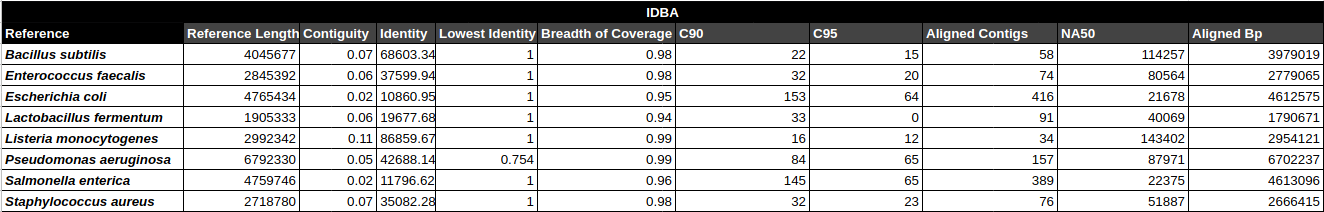
- Contiguity
- Identity (average)
- Identity (min)
- Breadth of coverage
- C90 & C95
- Number of aligned contigs
- Phred quality score per contig
Benchmark
| Assembly evaluation
C90 & C95
Number of contigs to cover at least 90% and 95% of the reference genome, respectively.
Contig Phread Quality Score
Contiguity
Longest percentage of the reference sequence assembled in a single contig.
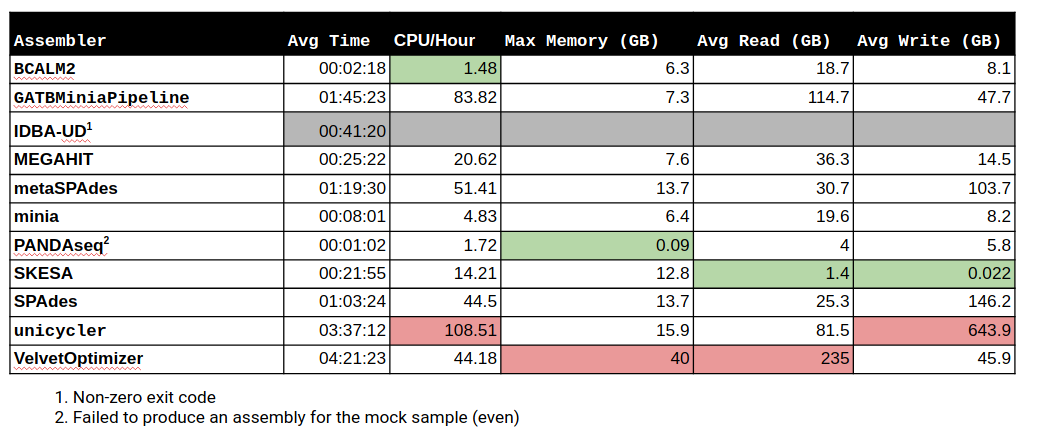
Benchmark
| Performace
Benchmark
| Mock sample (even)
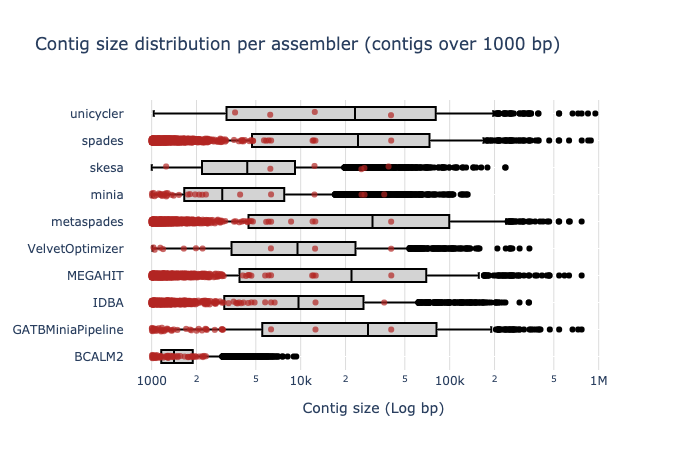
Benchmark
| Mock sample (even)
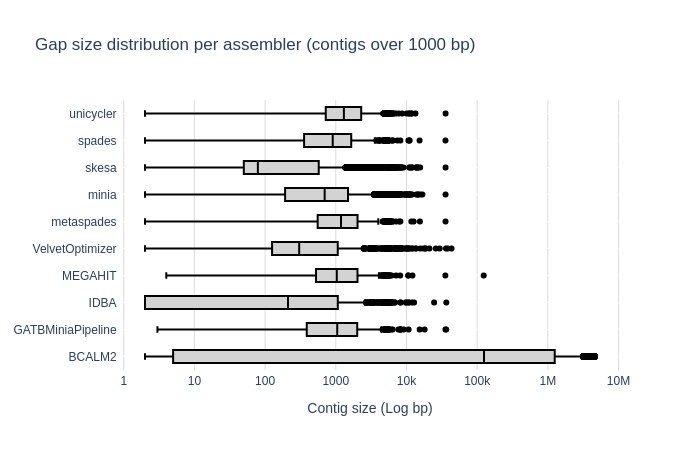
Benchmark
| Mock sample (even)
Benchmark
| Mock sample (even)
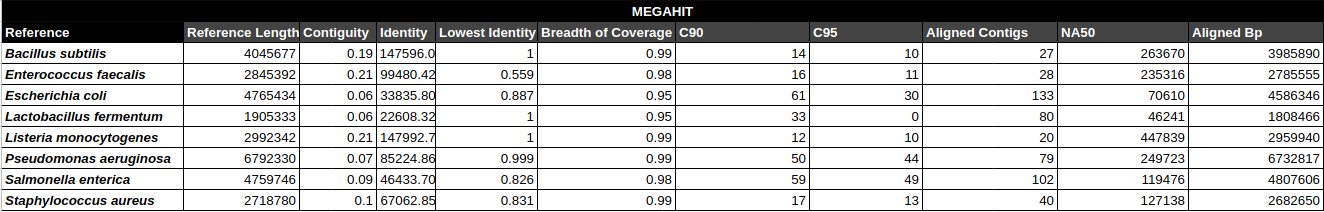
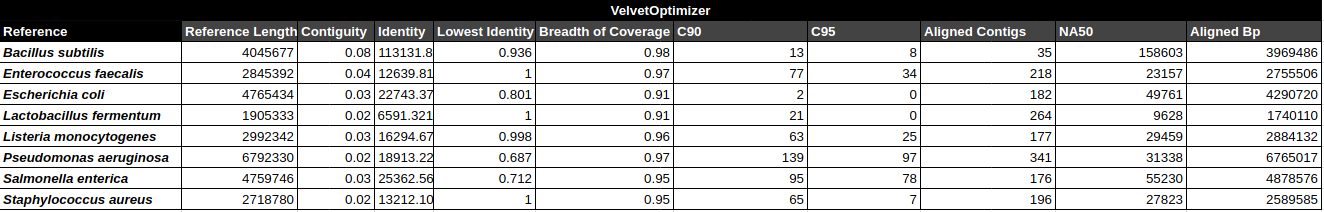
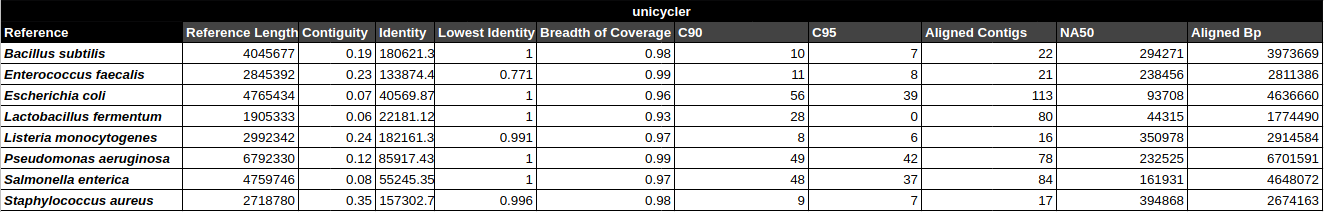
Bradth of coverage and number of contigs per Reference for each Assembler
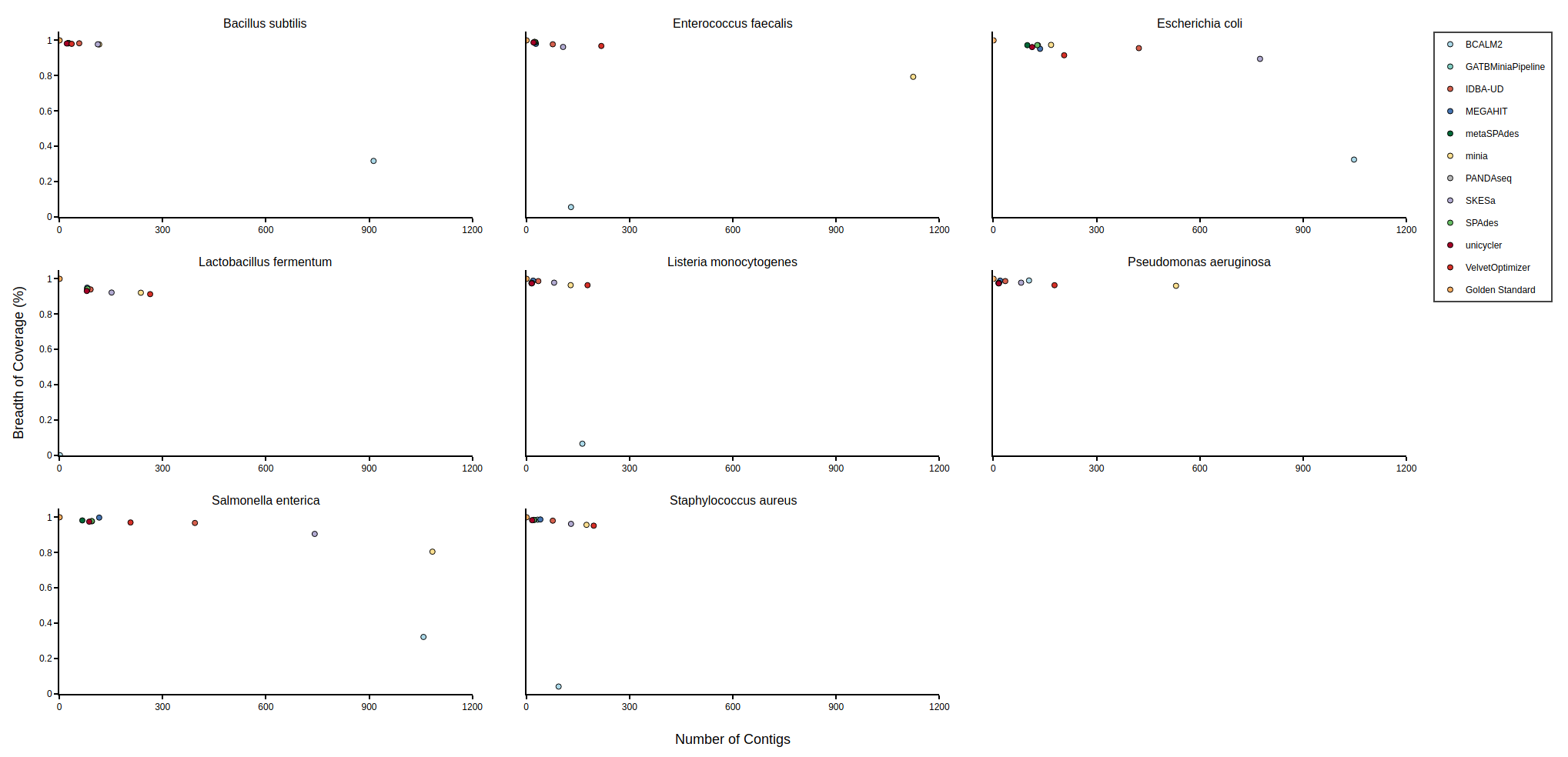
Benchmark
| Mock sample (even)
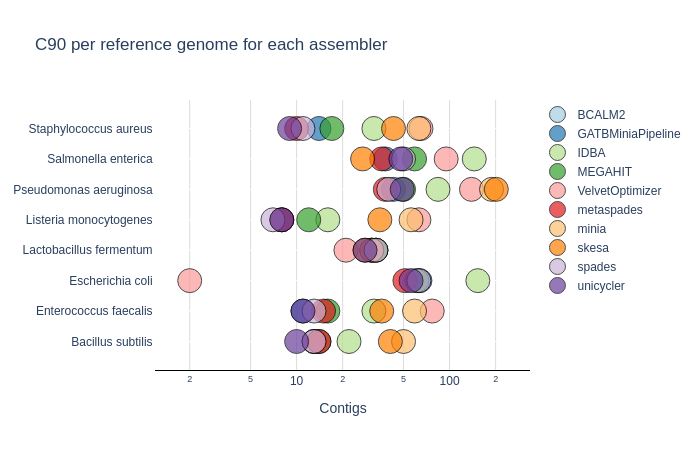
Benchmark
| Mock sample (even)
Contig Phred Quality Score for GATBMiniaPipeline's Pseudomonas aerugiona assemby
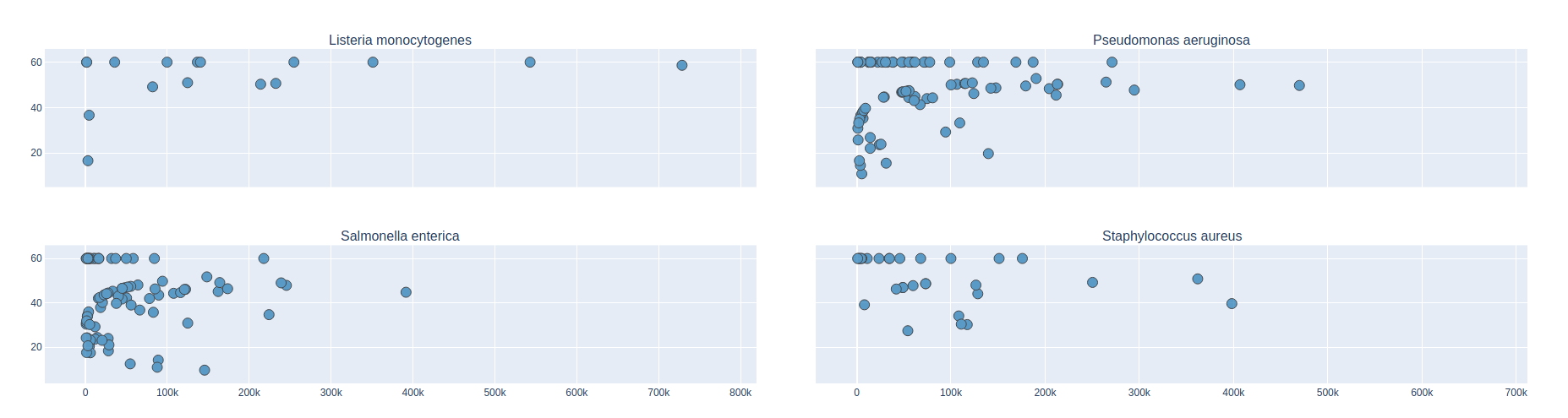


Contig Size
Phred Score
Benchmark
| Mock sample (even)
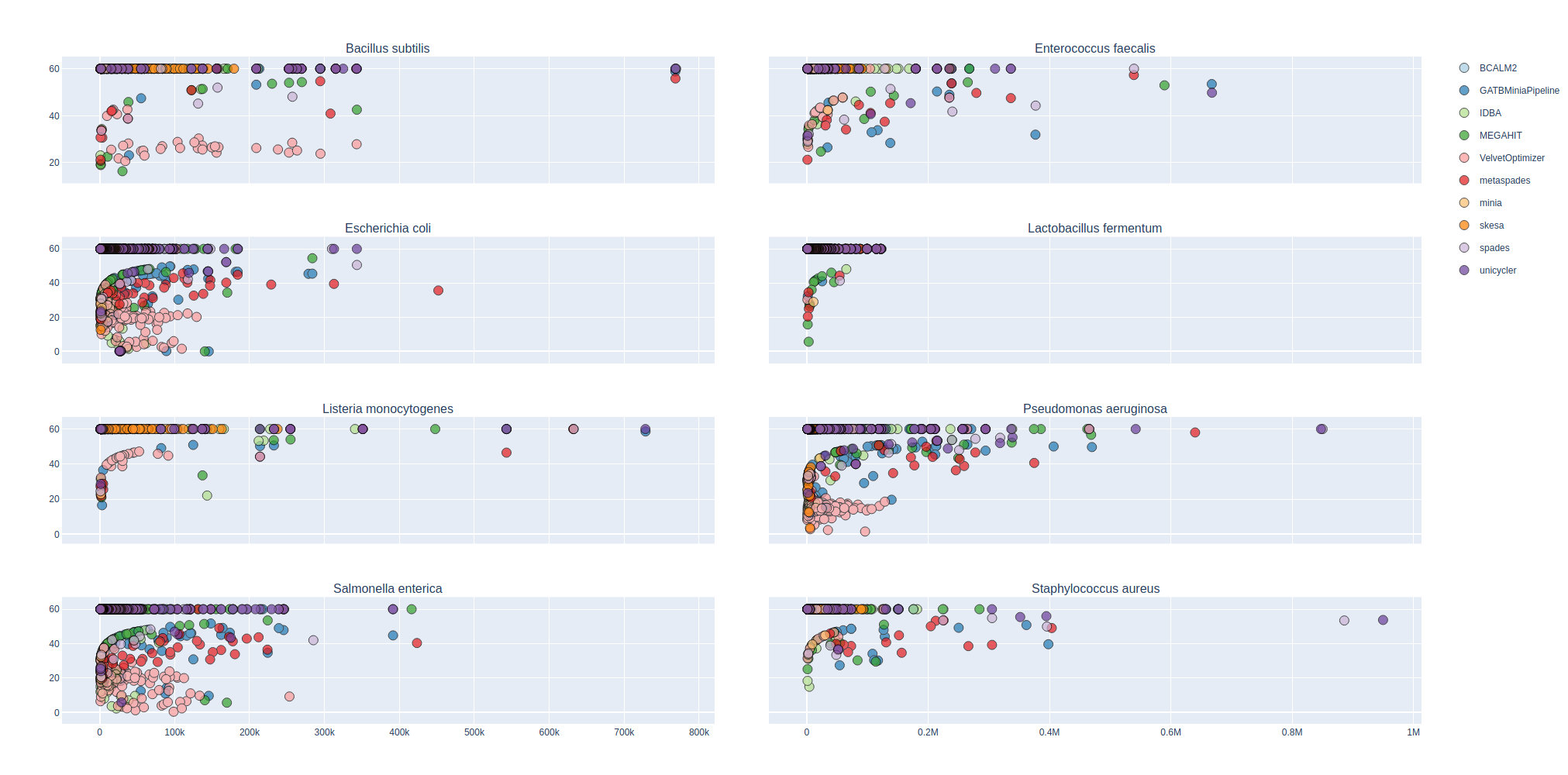
Contig Phred Quality Score per Reference for each Assembler

Data Structures & the SARS-CoV-2 Contextual Data Specification
Programa de Doutoramento do Centro Académico de Medicina de Lisboa
Inês Mendes
15th of September, 2020

PHA4GE
| Data structures workgroup
Standardized data structures and interchangable formats - critical to the development of an open software ecosystem.
Focus on the development, adaptation and standardization of data models for microbial sequence data, contextual metadata, results and workflow metrics to improve the transparency, interoperability and reproducibility of public health sequencing workflows.
Main Goal


PHA4GE
| SARS-CoV-2 Data Spec
SARS-CoV-2 contextual data specification that incorporates publicly available community standards, as well as additional fields and guidance appropriate for public health surveillance and analyses.
- Collection template and controlled vocabulary pick lists
- Reference guide
- Collection template SOP
- Data submission protocol (GISAID, NCBI, EMBL-EBI)
- JSON structure of PHA4GE specification
Resources

PHA4GE
| SARS-CoV-2 Data Spec

Future Work
Continue benchmark analysis for the mock semple (log distributed) and the real samples (Zymos community standards log and evenly distributed).
PHA4GE - Harmonization of tool outputs for the detection of antimicrobial registance genes (https://github.com/pha4ge/hAMRonization).
Web service for the interactive vizualization of Kraken's taxonomic composition reports.
Nothing else Meta - Reference indenpendent filtration of human reads from (meta)genomic datasets.
Special thanks to Diogo Silva, Bruno Gonçalves, Tiago Jesus, Pedro Vila-Cerqueria, Rafael Maria Mamede, João Carriço, John Rossen and Mário Ramirez.







Thank you for your attention

