Transparency in Bioinformatics


@ines_cim
cimendes
Inês Mendes
Instituto de Medicina Molecular João Lobo Antunes
Through the looking glass
of Data and Software
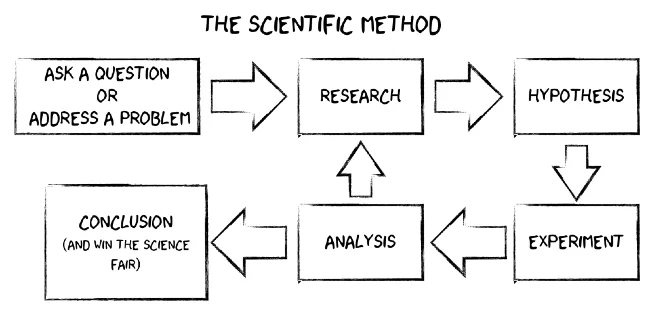
Can person X, with the same data and the same methodology, obtain the same conclusions as person Y?





Black
Box
Glass
Box
- Commercial/Freeware
- You get what it gives you
- Ready to use
- Stealth change
- Standalone
- Freeware
- You can "tailor"
- "Major" headache
- Visible change
- Dependencies
Reproducibility
| The needs


- Do I know what is happening?
- Is it reproducible?
- Is it shareable?

Reproducibility
| The needs
The needs:
- Analyze a large amount of sequence data routinely
- Some computationally intensive steps
- Constantly updating/adding software

Reproducibility
| The needs
Reproducibility
| The challenges
- No standard way of describing experiments, environments, (derived) data, and workflows.
- No transparency in creating environments and steps/methods to recreate analysis.
- The experimental nature of the research code and ecosystem makes it often hard to build.
- Unresolved or undocumented dependencies.
- Infrastructure for storage and distribution.
What is our role, as computational biologists, in addressing these challenges?
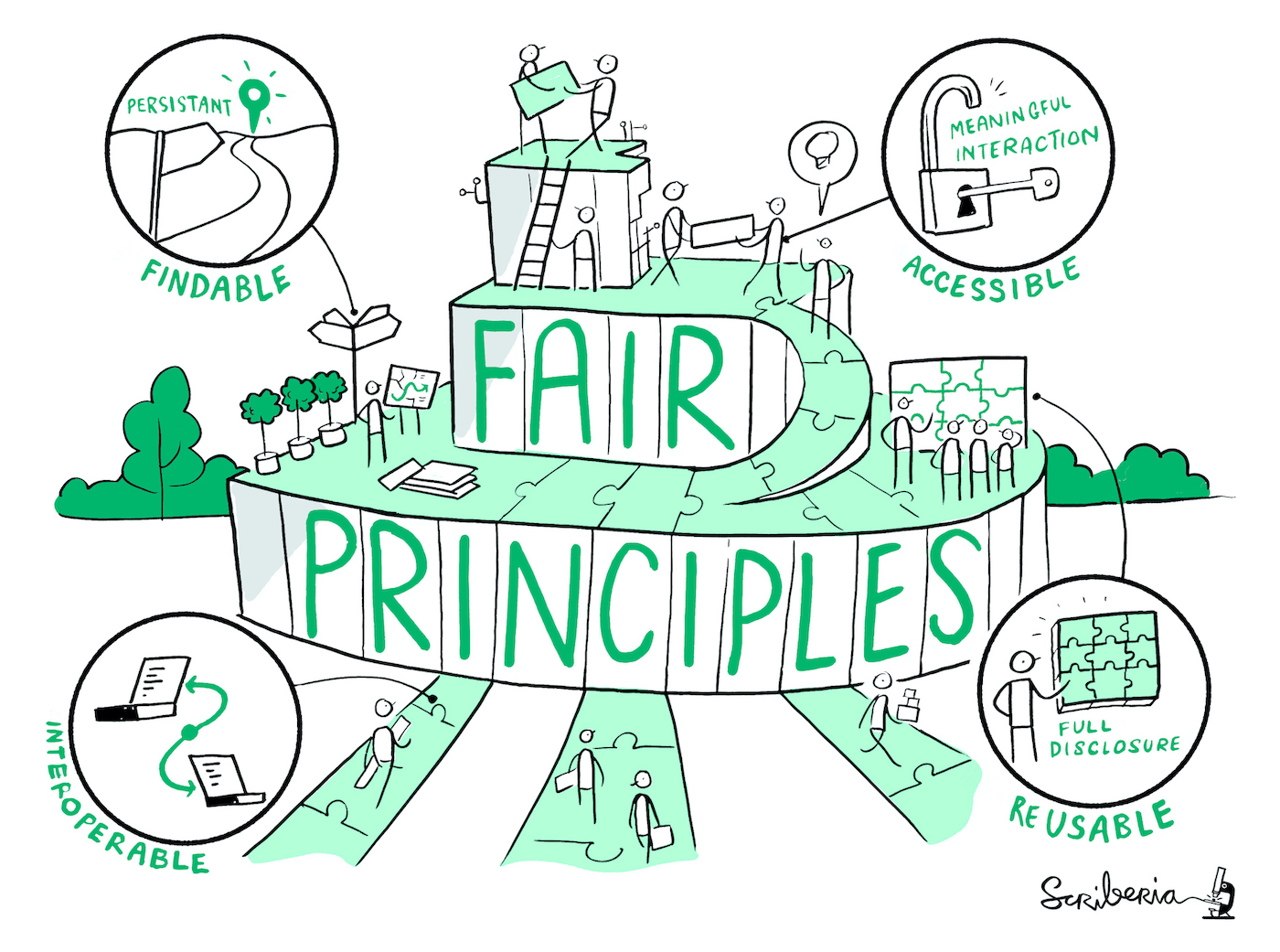
FAIR Data Principles
Findable, Accessible, Interoperable, Reusable
FAIR Principles
| Not just for data
https://doi.org/10.5281/zenodo.3332807
FAIR Principles
| Not just for data
https://doi.org/10.1038/sdata.2016.18
FAIR Principles
| Not just for data
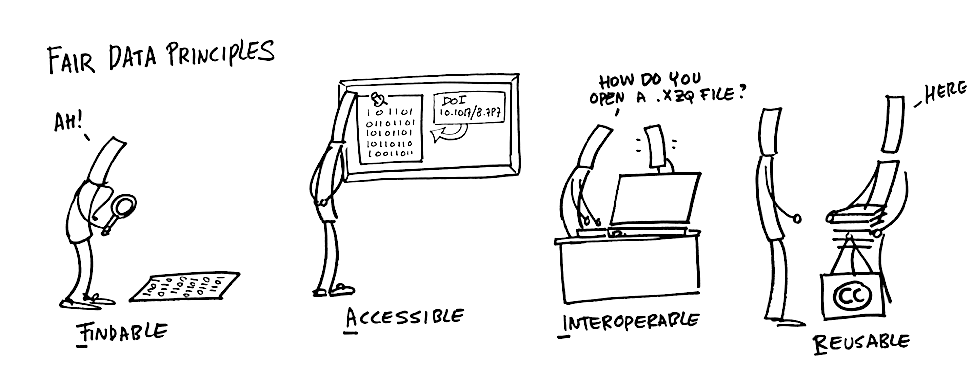
Findable: The first step in (re)using data is to find them! Descriptive metadata (information about the data such as keywords) are essential.
Accessible: Once the user finds the data and software they need to know how to access it. Data could be openly available but it is also possible that authentication and authorisation procedures are necessary.
Interoperable: Data needs to be integrated with other data and interoperate with applications or workflows.
Reusable: Data should be well-described so that they can be used, combined, and extended in different settings.
But software is not data.
- Everyone, everywhere


FAIR Principles
| Not just for data
The quality of the form of the software can be covered by FAIR data principles
- Code quality
- Maintainability
The quality of the functionality of the software goes beyond the FAIR principles:
- Correctness
- Security
- Efficiency
Form versus function of software
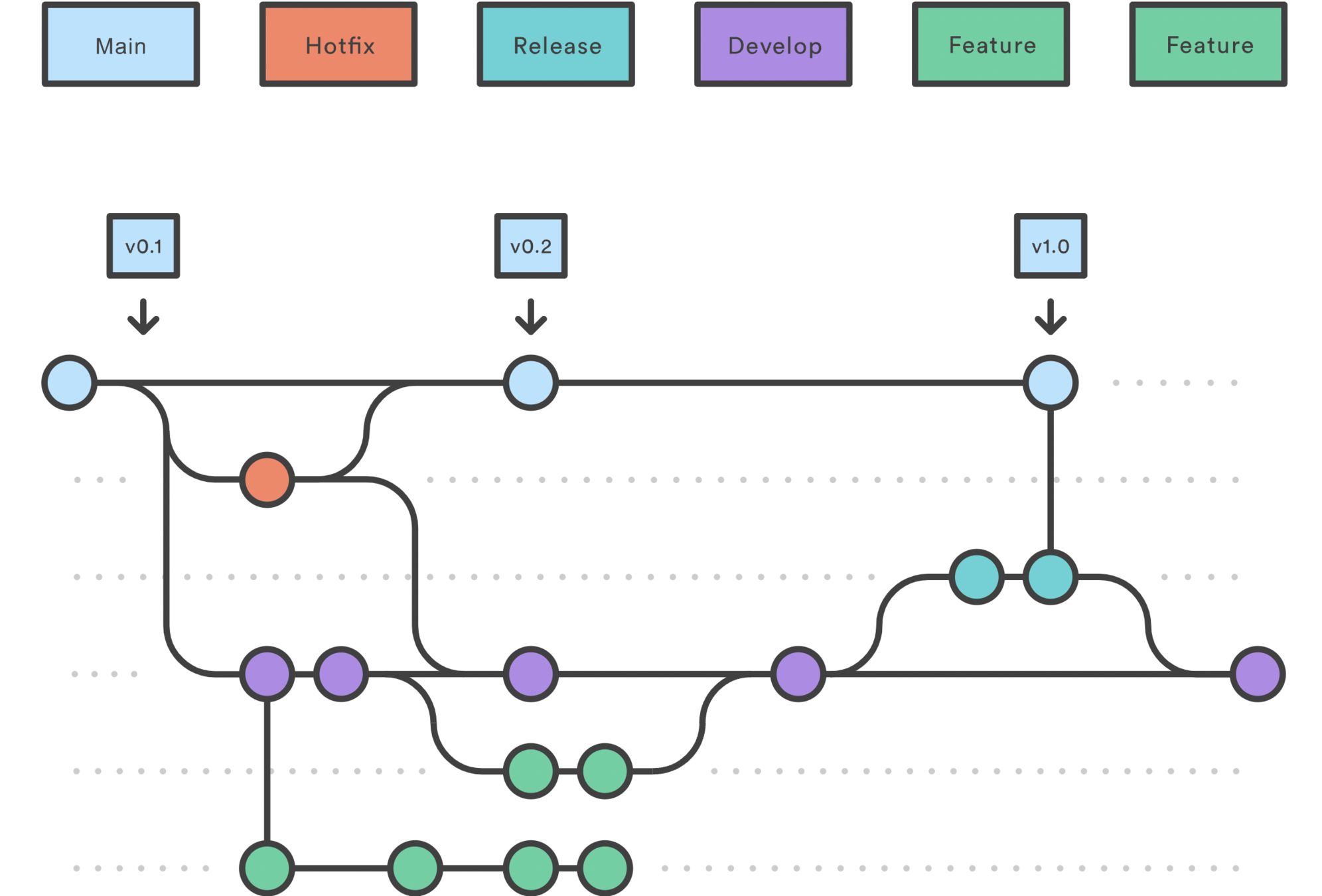
Version Control
Versioning, Collaboration and Accountability
Version Control
| What is it?
Version Control






| What is it?
Version Control
| What does it allow for?
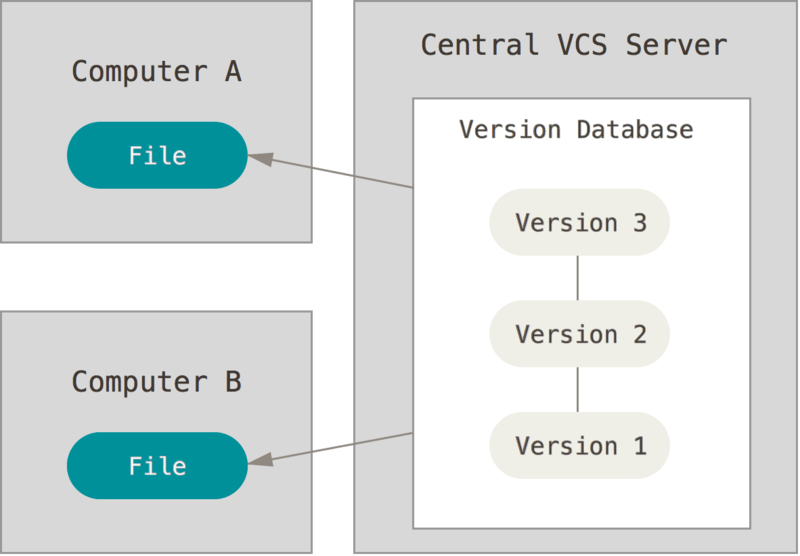
Collaboration: VCS (such as Git) was designed to solve the problem of multiple people working on the same code.
Storing versions properly: A stage of the project can be saved, through a commit, alongside a message. In the end, you only have one version on your disk of a project. Everything else is neatly packed up inside the VCS.
Auditabillity: VCS offers you an easy way to track "who made what change and when", allowing you to (1) debug more effectively, (2) track the reason that certain changes were made and (3) find the person who made a change.
Container Software
Reproducibility, Deployability
Software Containers
| What is it?
In the Paleolithic era:

Virtual Machines
“Bare Metal” Installation
Software Containers
| What is it?
In the Modern days:


Software Containers

Software Containers
Runs the same regardless of the environment.
A container image is a lightweight, stand-alone, executable package of a software that includes everything needed to run it:
- code
- runtime
- system tools
- system libraries
| What is it?

Software Containers
| What is it?
Host Hardware
Host Hardware
Container Engine
Host OS
Host OS
Hypervisor
Guest OS
Guest OS
App
App
Guest OS
VM1
VM2
App
App
App
App
Virtual Machines
Containers
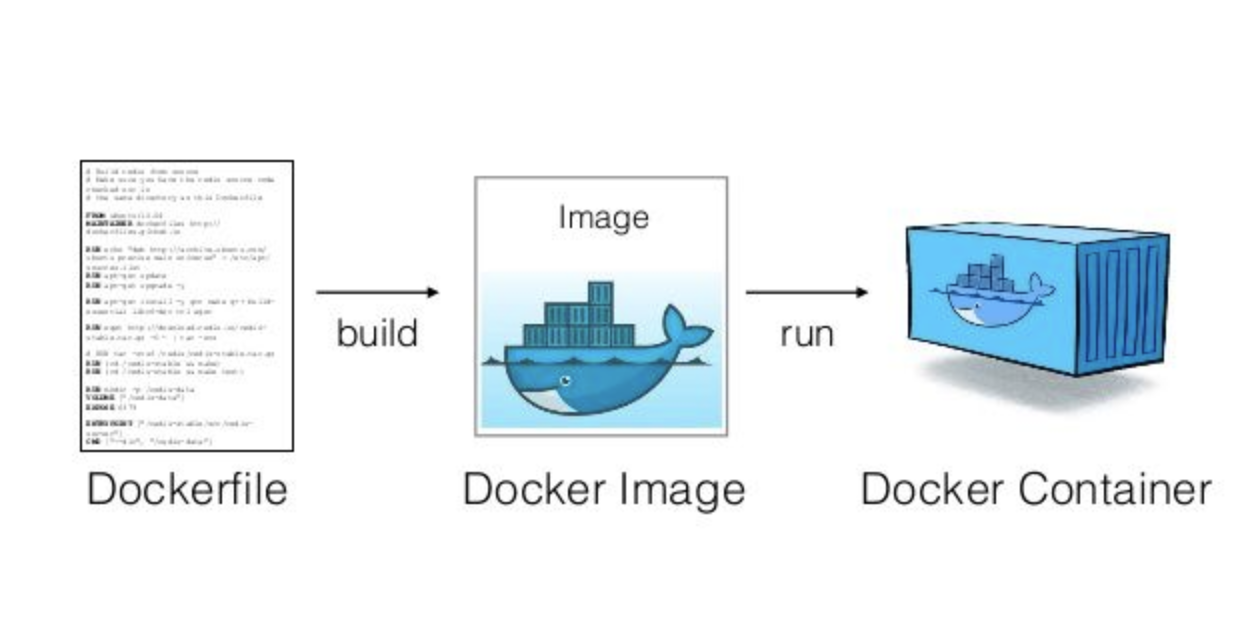


Docker Hub
build
pull & run
host
push
Docker
| The ecosysthem
Workflow Managers
Reproducibility, Scalability, Sharability
The game changing combination of workflow managers and containers:
- Portability
- Reproducible
- Scalability
- Multi-scale containerization
- Native cloud support







Reproducibility
| The motivation
Workflows in the Modern era:
Workflow Managers
| What is it?



Enables scalable and reproducible scientific workflows. It simplifies the deployment of complex parallel and reactive workflows.
Reactive workflow framework
Create pipelines with asynchronous (and implicitly parallelized) data streams
Programing DSL
Has its own language for building a pipeline
Containerized
Out of the box integration with containers engines (Docker, Singularity, Shifter)
The creation of workflow pipelines was designed for bioinformaticians familiar with programming.
It's execution is for everyone.
Workflow Managers
| What is it?
- Automatic management of temporary input/output directory/files
- No need for custom handling of concurrency (parallelization)
- A single pipeline can support any scripting language (Bash, Python, Perl, R...)
- Every process (task) can be run in a container
- It's portability allows for the same pipeline to run on a laptop, server, cluster, etc
- Checkpoints and resume functionality
- Host pipeline on GitHub and run remotely




Monolithic pipelines


Need to change often
Siloed tool containers
Don't do much by themselves
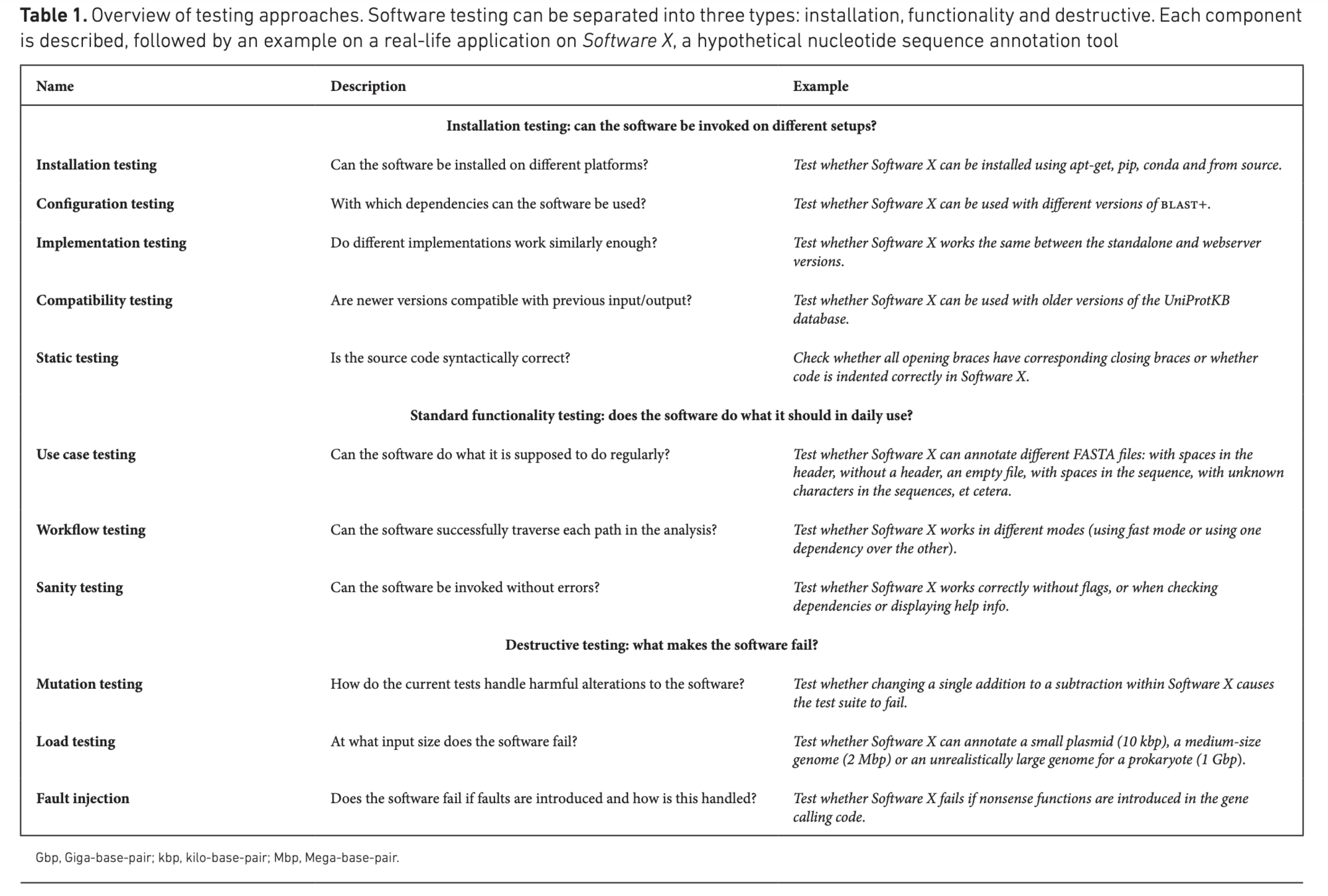
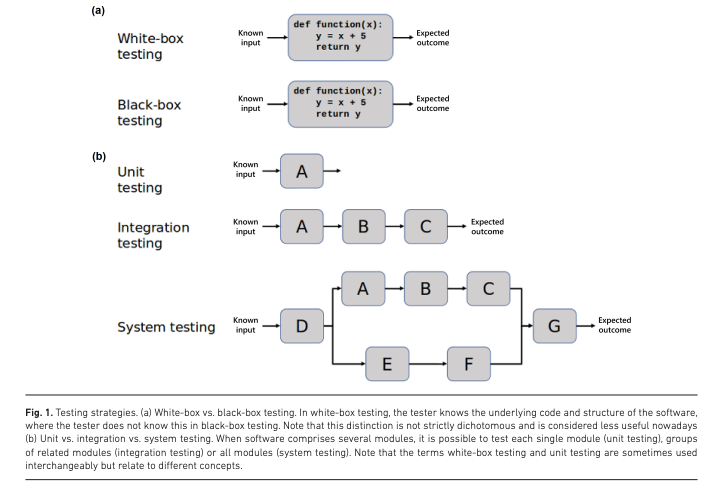
Software Testing
The final frontier
Code Development
| Approaches
Design
Code
Test
Test
Code
Refactor
Traditional Technique Approach
Test Driven Development Approach
In the field of microbial bioinformatics, good software engineering practices are not yet widely adopted. (...) This paper serves as a resource that could help microbial bioinformaticians get started with software testing if they have not had formal training.


Recommendation #1
Establish software needs and testing goals
Recommendation #1
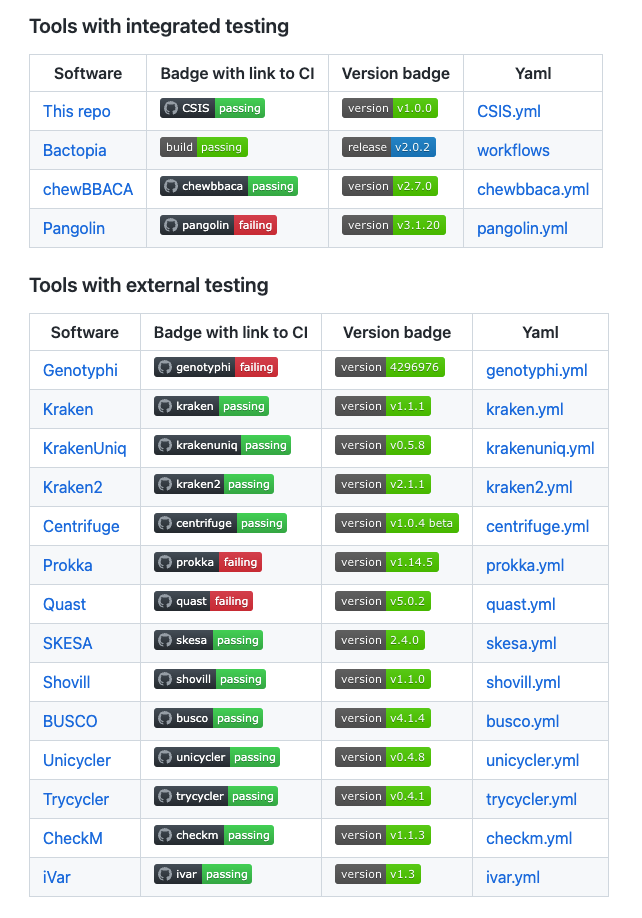
The CSIS repository aims to promote the uptake of testing practices and engage the community in its adoption for public health.
This repository is an open-source project that gathers guidance, guidelines and examples for software testing for microbial bioinformatics researchers.
A proof of concept that the adoption of new standards can be crowdsourced.
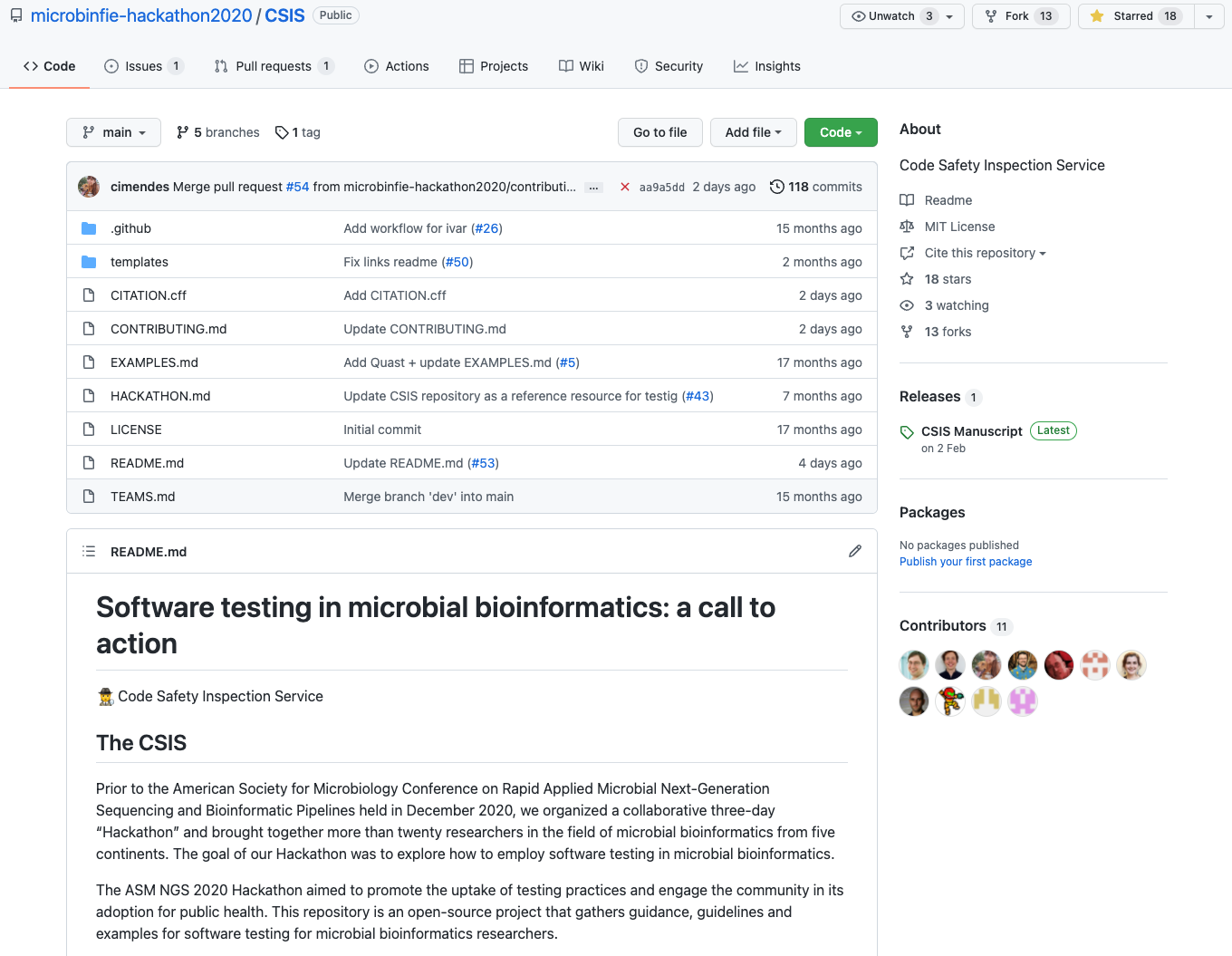
Recommendation #2
Input test files: the good, the bad, and the ugly
Include test files with known expected outcomes for a successful run.
Include files or other inputs on which the tool is expected to fail.
Recommendation #3
Use an established framework to implement testing

unittest (https://docs.python.org/3/library/unittest.html)
pytest (https://docs.pytest.org/en/stable/)

(https://testthat.r-lib.org/) testthat
Recommendation #3

Recommendation #4
Testing is good, automated testing is better


Recommendation #5
Ensure portability by testing on several platforms
Recommendation #6
Showcase the tests



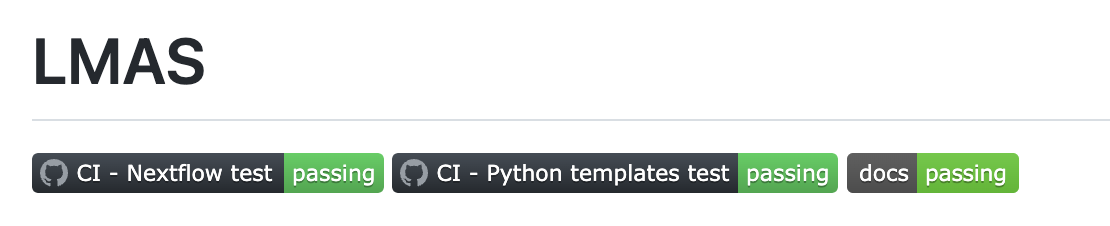
Recommendation #7
Encourage others to test your software!



Try it out!





Thank you for your attention
Special thanks to Pedro Vila-Cerqueira, Rafael Mamede and Mário Ramirez.
FCT PhD Grants SFRH/BD/129483/2017
COVID/BD/152618/2022

MRamirez Lab, iMM
2019

