Inês Mendes
MRamirez Lab - iMM

@ines_cim
cimendes
Metagenomic Assembly
Towards Accreditation in Metagenomics for Clinical Microbiology
Advisor Comittee Meeting
14 July 2021
Microbial (meta)genomics
Shotgun Sequencing Data
- Direct Read Classification Methods
- Assembly-Based Methods
- Marker Gene Detection
- Taxonomical Assignment
- Abundance estimation
- Sequence binning
- Abundance estimation
- Computational expensive
- Lacks context
- De novo metagenomic assemby
- De novo metagenomic co-assemby
- Consensus metagenomic assemby
- Provides contextual information
- Normalizes information
Microbial (meta)genomics
| Assembly
The assembly methods provide longer sequences that are more informative than shorter sequencing data and can provide a more complete picture of the microbial community in a given sample.

Assembly
consensus
de novo
OLC
de Bruijn graph
Microbial (meta)genomics
| Assembly
- Results are highly dependent on the tools chosen for the analysis - Lack of standardization and proper benchmark.
Major issues
- Highlights the potential and the limitations of shotgun metagenomics as a diagnostic tool - Lack of reproducibility
Reads
Contigs
Genomes
Microbial (meta)genomics
| Assembly
- State-of-the-art software assessment for short-read metagenomic de novo assembly (Task 1)
Main goals
- Evaluation of the influence the choice of assembler has on quality, reproducibility and resource requirements for metagenomic analyisis
- Development of a reproducible, user-friendly workflow for the assessment of assembly success (Task 2)
Reads
Contigs
Genomes
Microbial (meta)genomics
| Assembly
- Limited to short-read paired-end sequencing data
Main limitations
Reads
Contigs
Genomes
- Limited to de novo assembly software
- Lack of flexibility for the introduction of new tools
7 months ago...
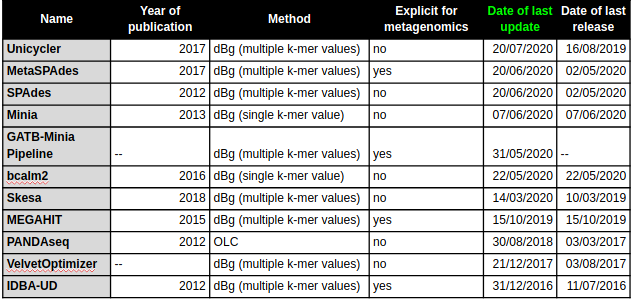
Compiled a collection of de novo assembly tools, including Overlap, Layout and Consensus (OLC) and De Bruijn graph (dBg) assembly algorithms, with both single k-mer and multiple k-mer value approaches, and hybrid assemblers. The collection includes both genomic and metagenomic assemblers.
7 months ago...
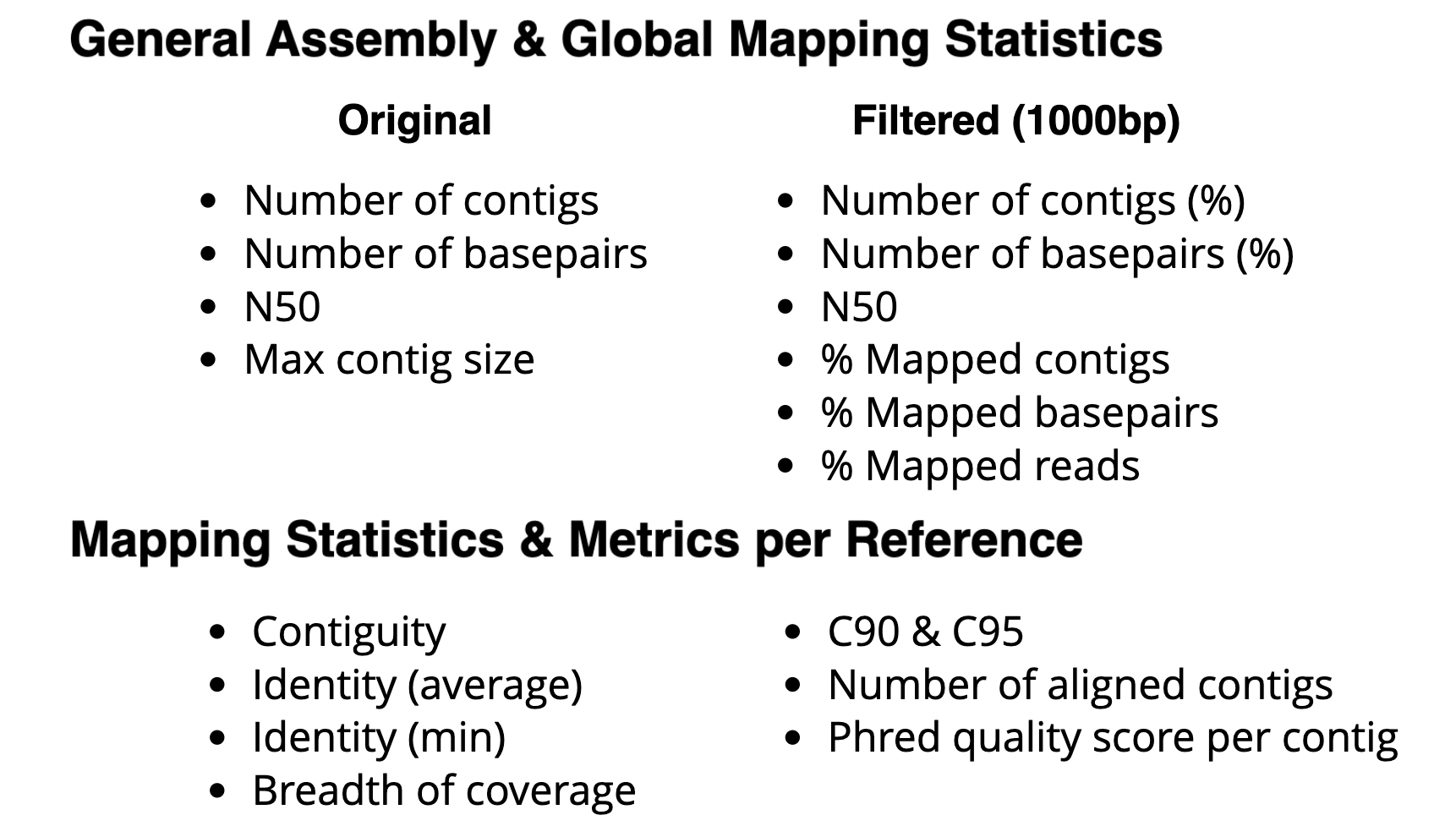
Initial development of global and reference specific performance metrics for the assessment of de novo assembly quality.
7 months ago...
https://github.com/cimendes/metagenomic-assembler-comparison
Reference Dataset (Complete Bacterial Genomes)
In silico mock sample (even)
In silico mock sample (log)
Zymos standard (even)
Zymos standard (log)
3.7 M read pairs
8.8 M read pairs
47.8 M read pairs
Basic Assembly Workflow
Assembly Quality Assessment

https://github.com/cimendes/LMAS
https://lmas.readthedocs.io/

LMAS
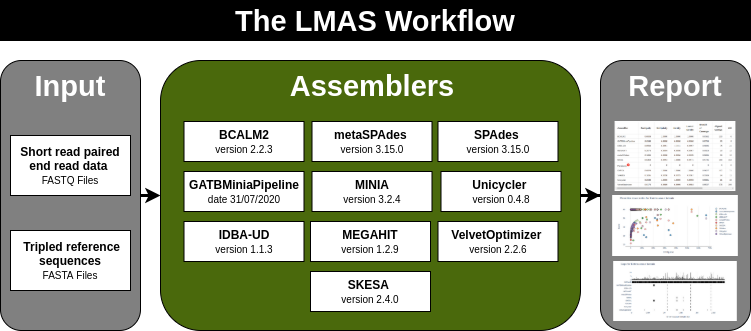
| Last Metagenomic Assembler Standing
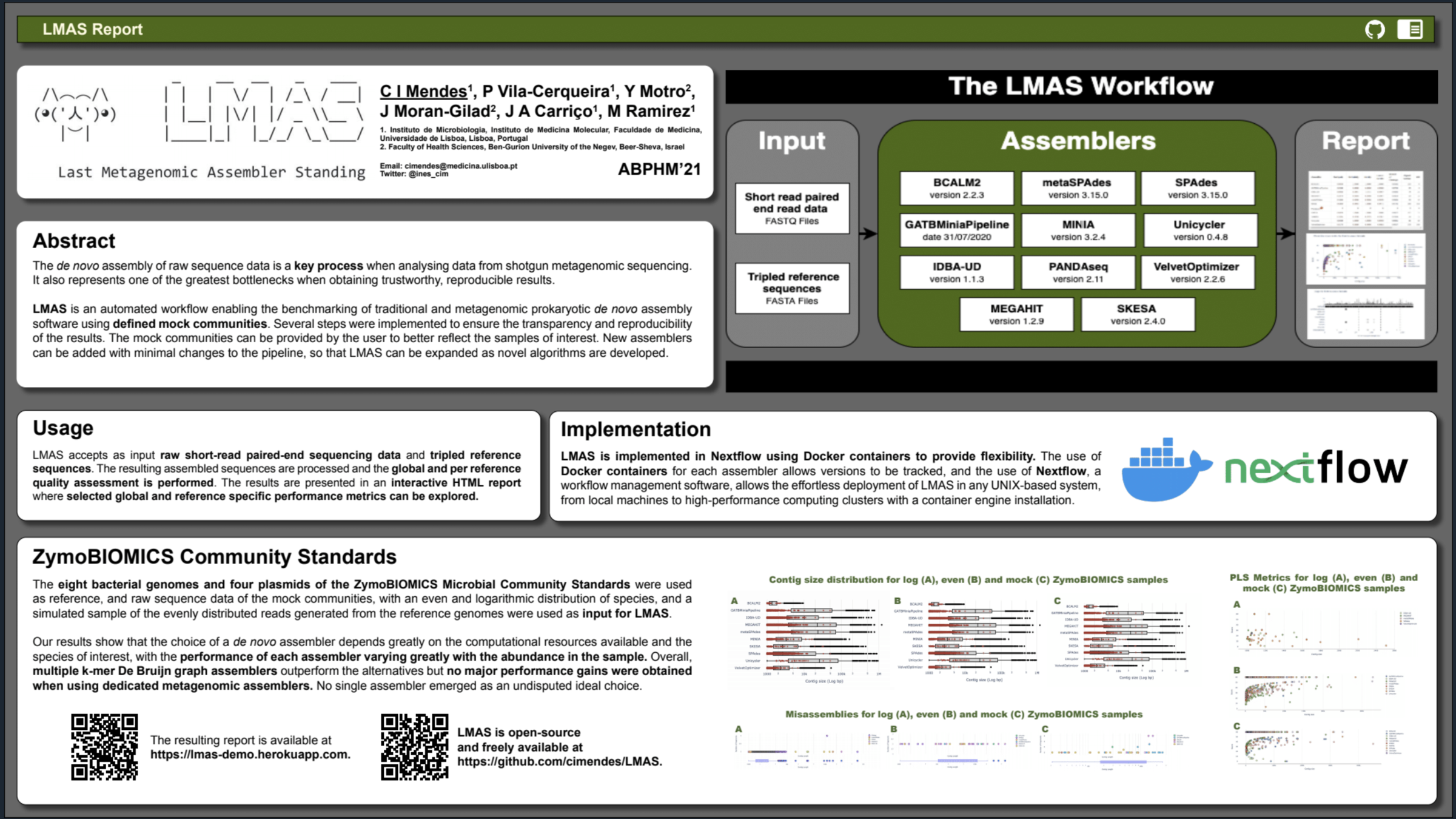
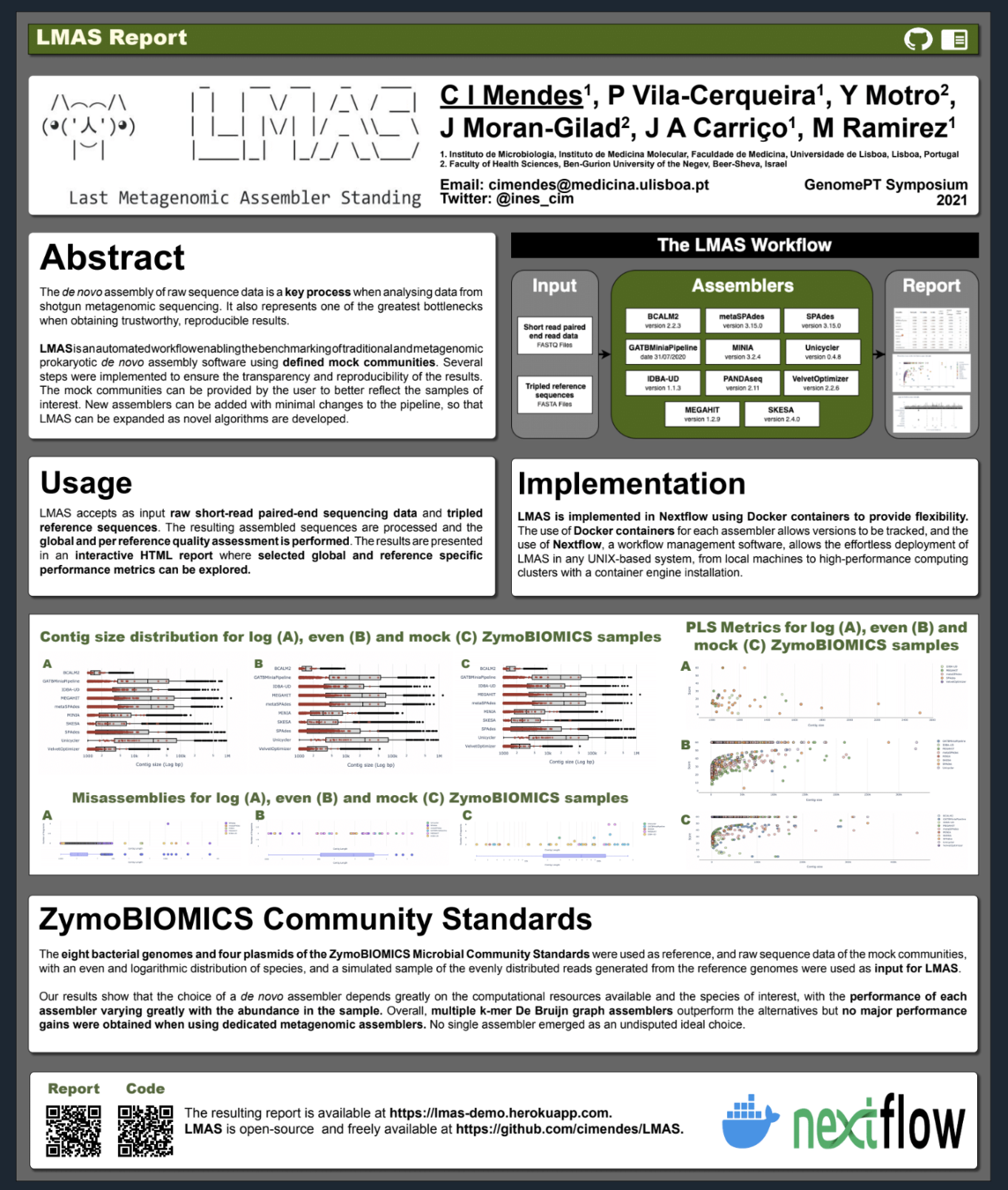
Automated workflow enabling the benchmarking of genomic and metagenomic prokaryotic de novo assembly software using defined mock communities.
LMAS
| Last Metagenomic Assembler Standing






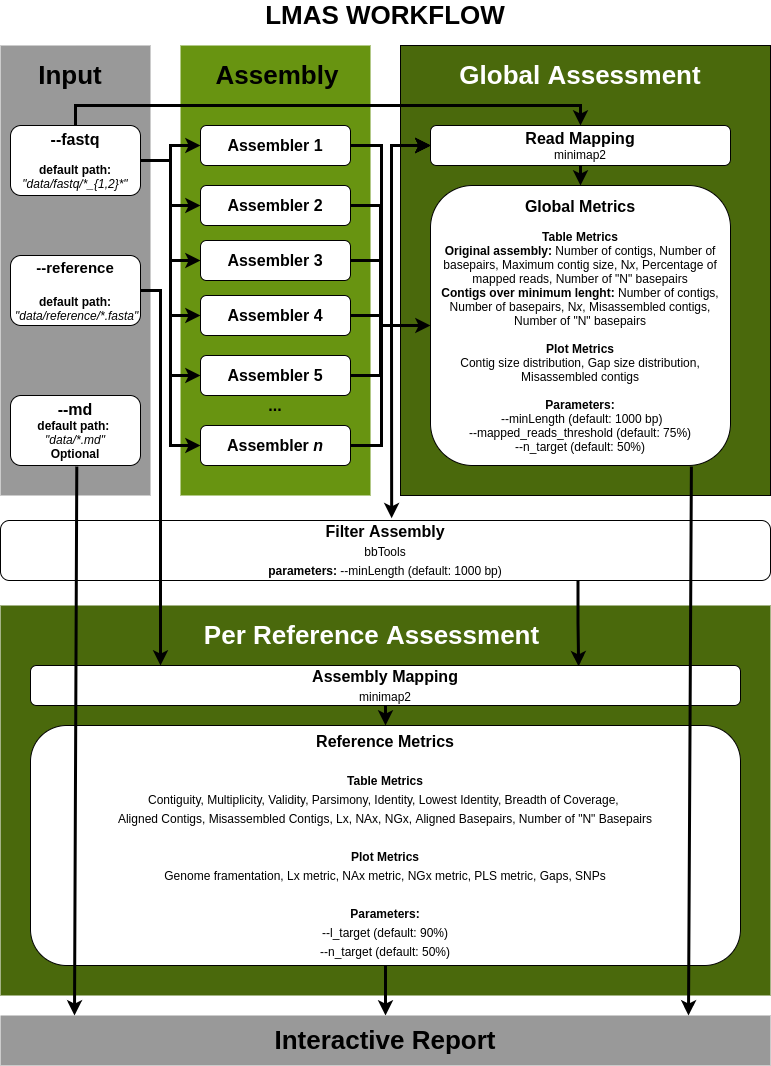



The input data is assembled in parallel by the set of genomic and metagenomic de novo assemblers in LMAS.
The global and per reference metrics are grouped in the interactive LMAS report for exploration.
The resulting assembled sequences are processed and assembly quality metrics are computed.
LMAS
| Last Metagenomic Assembler Standing
LMAS
| Last Metagenomic Assembler Standing
LMAS requires a Nextflow installation (version ≥ 21.04.1), requiring BASH and Java 8 (or higher).
All components of LMAS are executed in containers.
Continuous integration of the python templates for the quality assessment of assemblies is performed with pytest and GitHub Actions.
LMAS can be installed through Github.
LMAS
| Assembly Quality Metrics
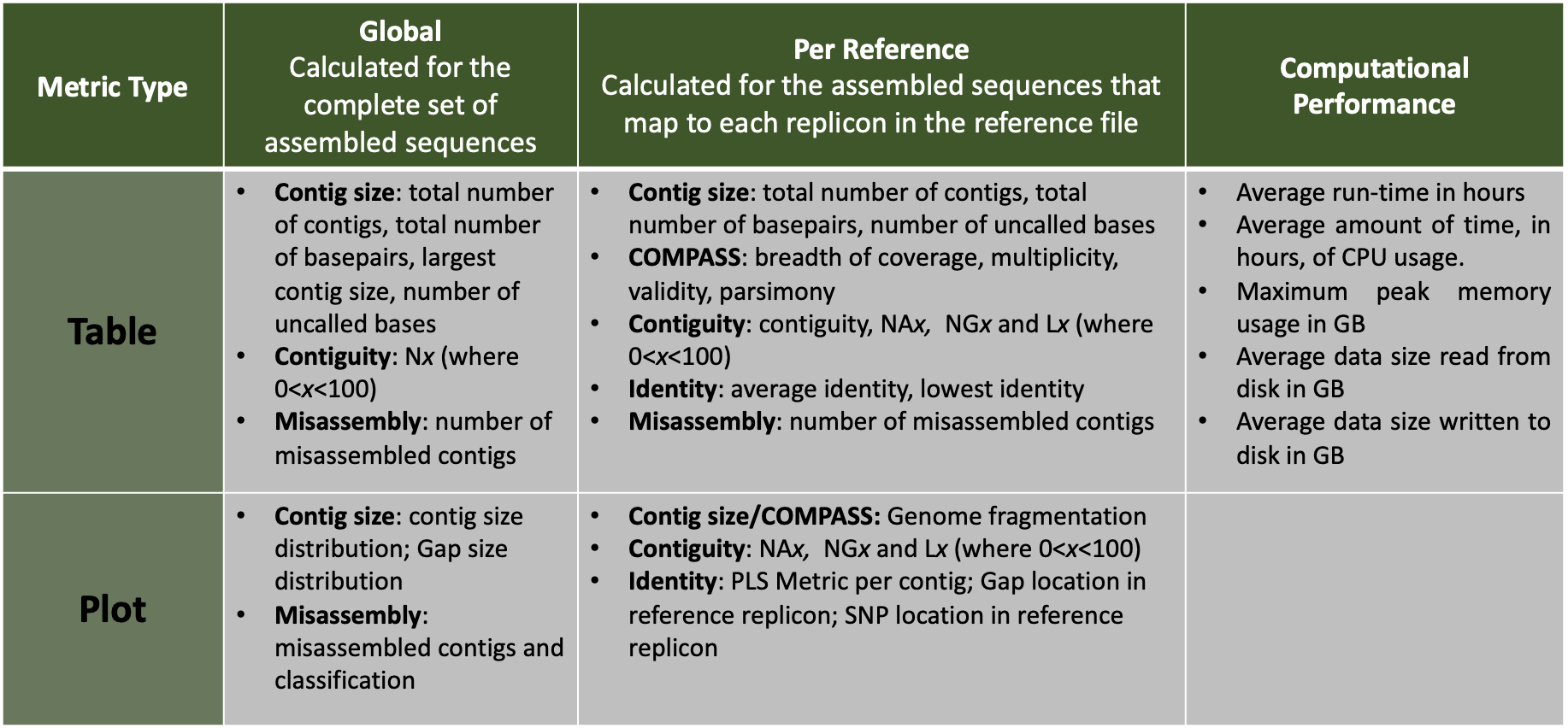
The tabular presentation allows direct comparison of exact values between assemblies, and the interactive plots allow for an intuitive overview and easy exploration of results.
LMAS
| Report
LMAS
| Report
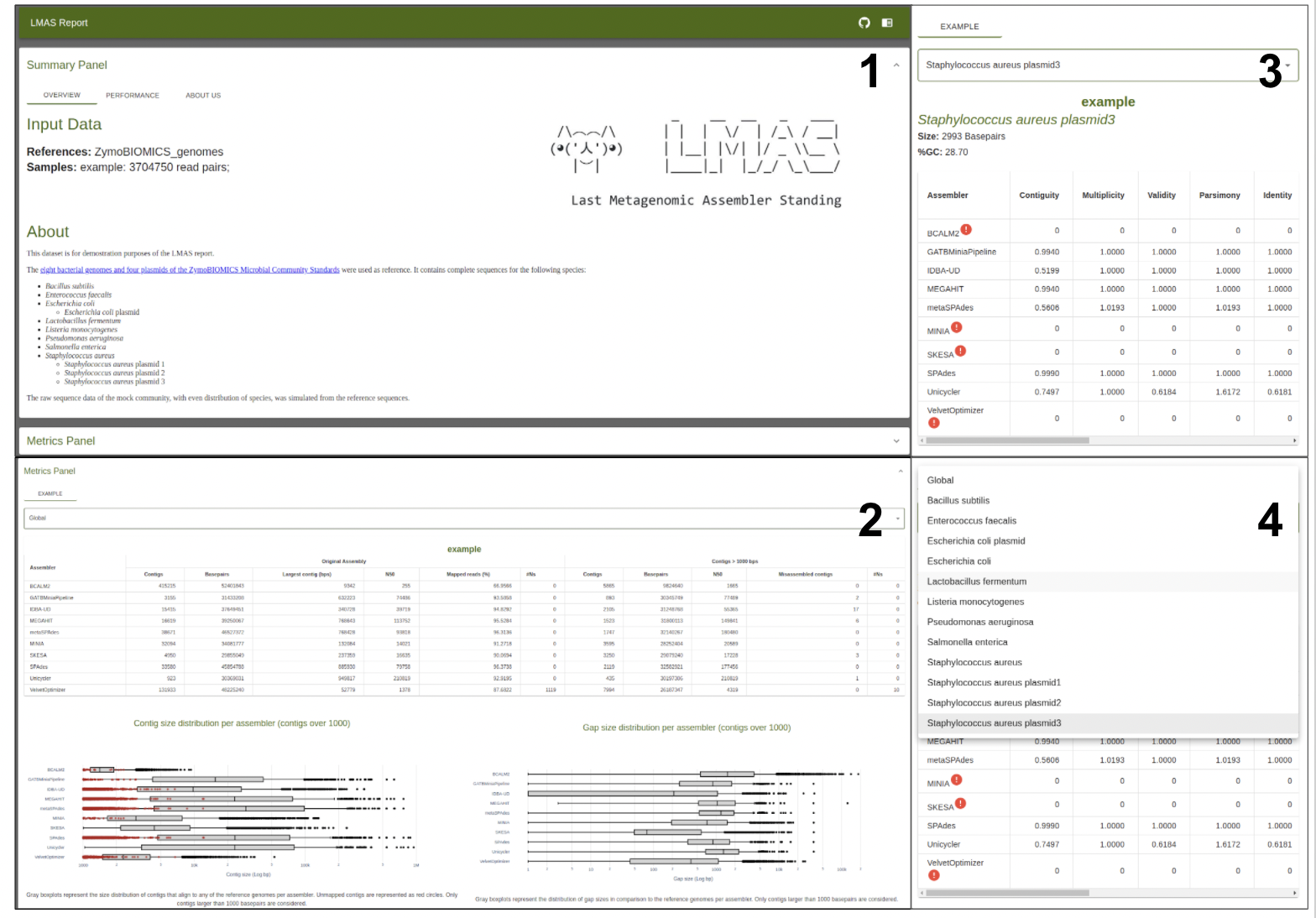
The results are presented in an interactive HTML composed of two main panels:
- a top summary panel with information on input samples and the LMAS execution,
- a bottom panel where selected global and reference specific performance metrics can be explored for each sample.
The JavaScript source code for the interactive report comes bundled with LMAS but is freely available at https://github.com/cimendes/lmas_report.

- Eight bacterial genomes
- Four plasmids
- Even and logarithmic distribution of species
- Simulated samples (with and without error) - InSilicoSeq
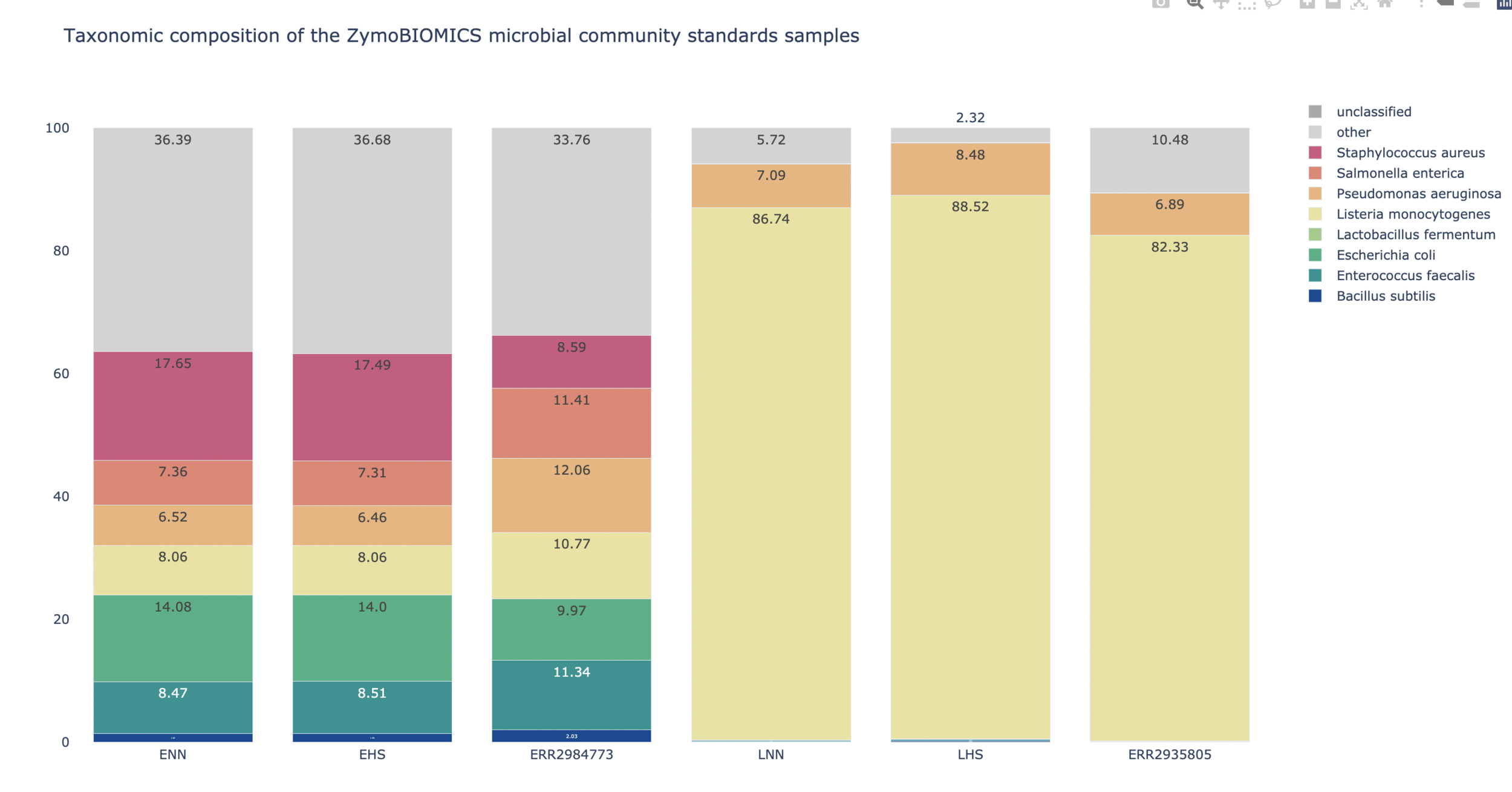
| Sample | Distribution | Error Model | Read Pairs (M) |
|---|---|---|---|
| ENN | Even | None | 8.6 |
| EHS | Even | Illumina HiSeq | 8.6 |
| ERR2984773 | Even | Real Sample | 8.6 |
| LNN | Log | None | 47.5 |
| LHS | Log | Illumina HiSeq | 47.5 |
| ERR2935805 | Log | Real Sample | 47.5 |
LMAS
| ZymoBIOMICS Microbial Community Standards
LMAS
| ZymoBIOMICS Microbial Community Standards
LMAS
| ZymoBIOMICS Microbial Community Standards
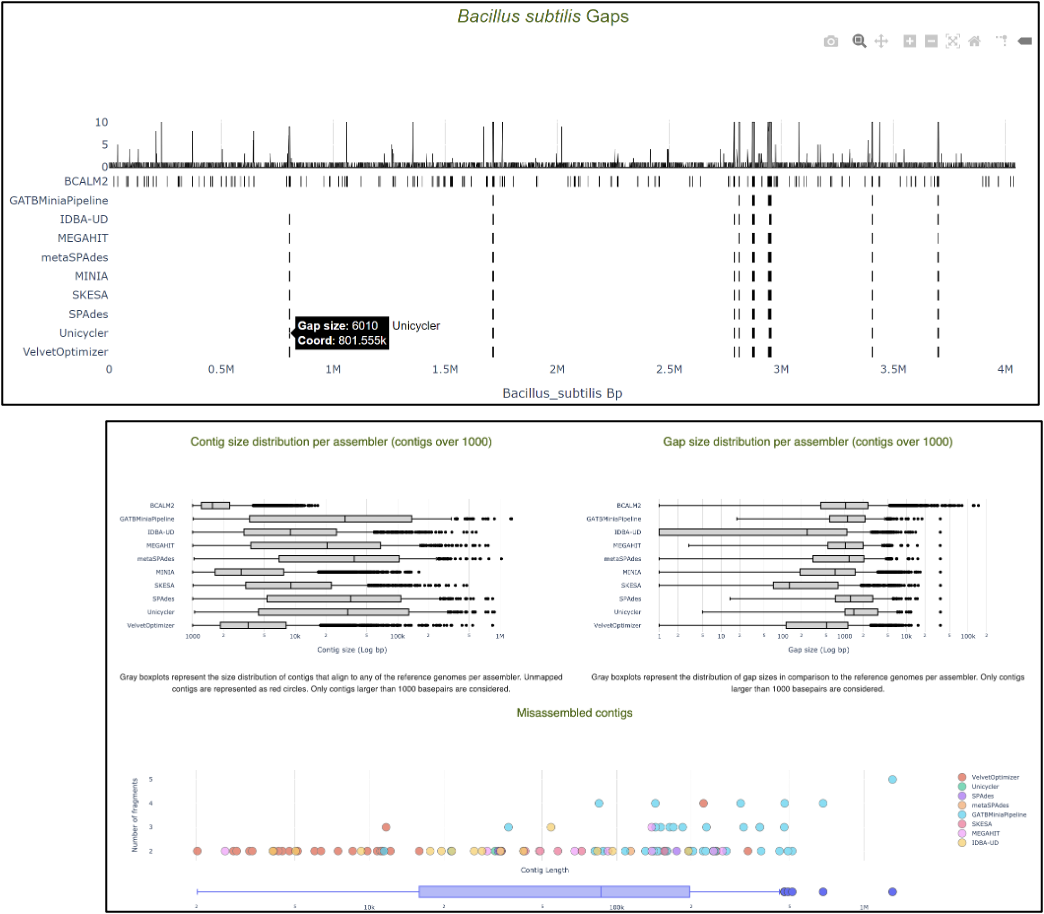
- Some assemblers perform poorly
- Resource usage is different
- Assembler performance is influenced by species abundance and sample composition
- Certain genomic regions are problematic for all assemblers
-
No one size fits all assembler!
LMAS
| 3 LMAS runs
https://github.com/cimendes/LMAS_Zymos_Resuls



LMAS
| 3 LMAS runs
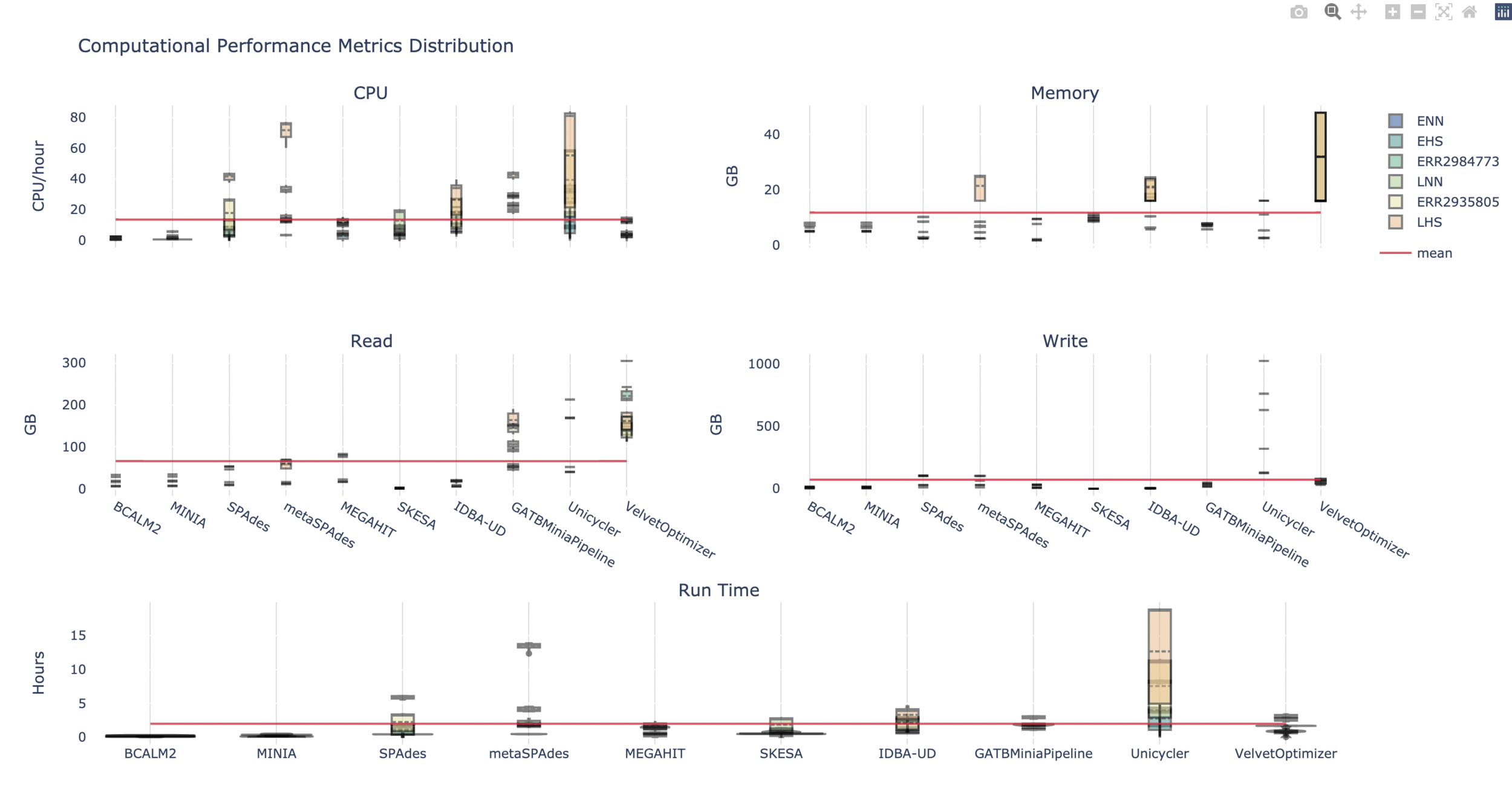
There’s a disparity in usage for the evenly and logarithmically distributed samples, with the latter having more resource-intensive requirements. The resource usage also varied greatly by assembler, with multiple k-mer dBG having overall higher resource usage.
LMAS
| 3 LMAS runs
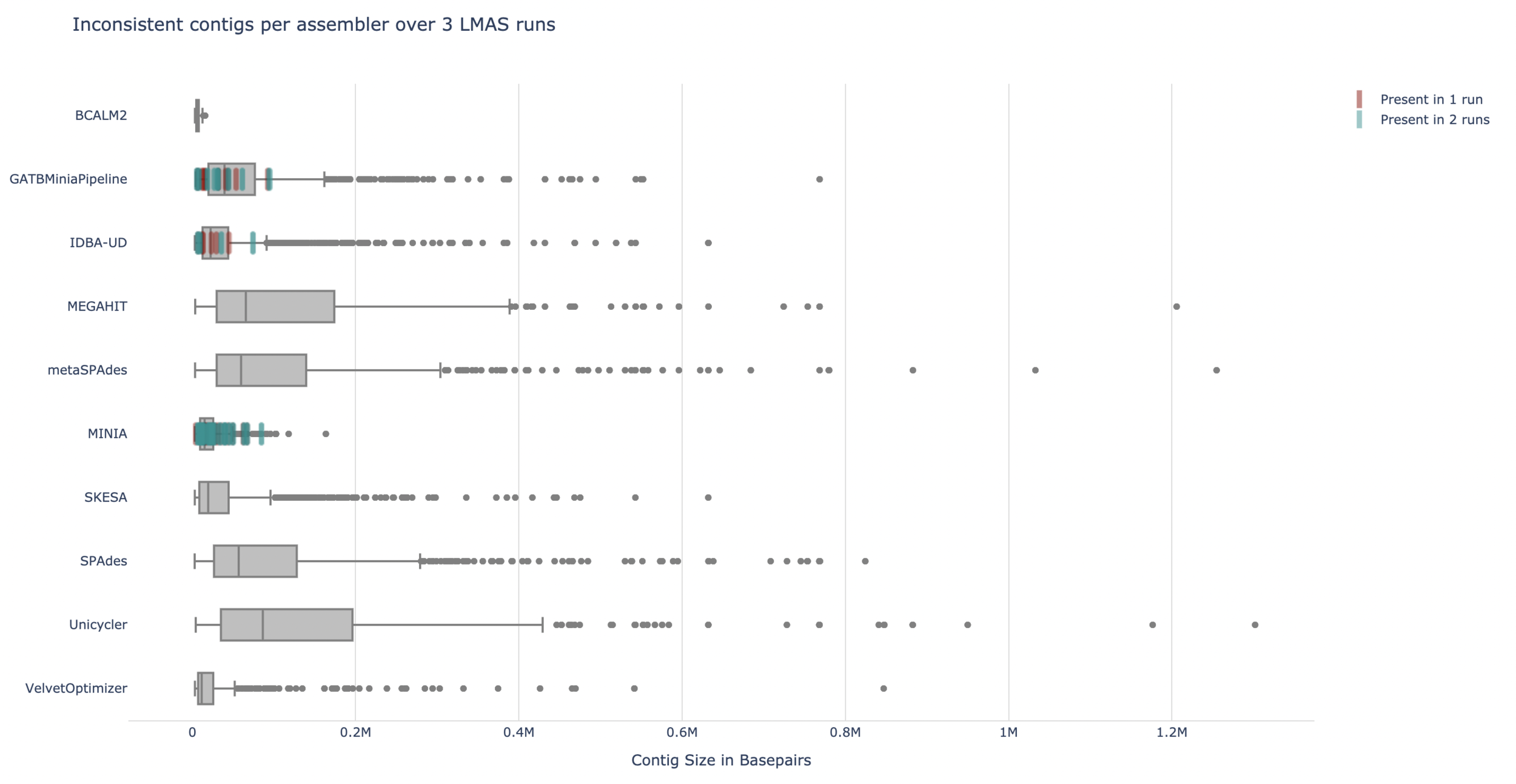
Only GATBMiniaPipeline, IDBA-UD and Minia assemblers produced inconsistent contigs (0.22%, 0.04% and 0.12% of the total contigs produced by the assemblers, respectively)
LMAS
| 3 LMAS runs
- The logarithmically distributed samples showed greater variation than the evenly distributed counterparts, and the real samples showed greater variation than the mock samples.
- The greatest difference observed was in the total basepairs produced by the assemblers but it didn’t impact the contiguity of the assemblies (N50) or the maximum contig size obtained.
- After filtering for the minimum contig length of 1000 basepairs, the effect of the variation in assembly length is mitigated, with all assemblers showing increased robustness in the number of contigs and total basepairs assembled.
LMAS
| Know issues
Generate mock dataset with matching read number and sample abundance distribution https://github.com/HadrienG/InSilicoSeq/issues/208
LMAS
| Know issues
LMAS
| Nice to have
Allow for long and hybrid assembly https://github.com/cimendes/LMAS/tree/dev_long_read


LMAS
| Nice to have
Allow for long and hybrid assembly https://github.com/cimendes/LMAS/tree/dev_long_read


Missing Points
- Manuscript Status
- Results Overview
- High priority tasks
- Lower priority tasks