dengue virus genotyping from amplicon and shotgun metagenomic sequencing

Micro Binfie Virtual Conference 2020
C I Mendes • U Lisboa
Twitter: @ines_cim
May 6th, 2020
Micro Binfie Virtual Conference 2020
Micro Binfie Virtual Conference 2020


Nabil made me do it....
Emma, I'll get you!
dengue virus genotyping from amplicon and shotgun metagenomic sequencing

Micro Binfie Virtual Conference 2020
C I Mendes • U Lisboa
Twitter: @ines_cim
May 6th, 2020
The dengue virus




| Transmission & Disease
The dengue virus
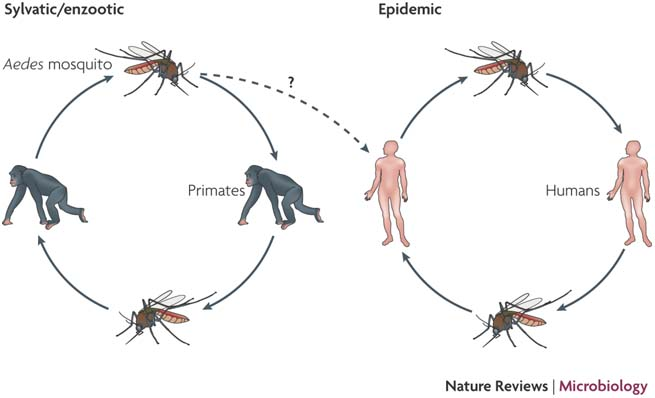
doi:10.1038/nrmicro1690
| Transmission & Disease
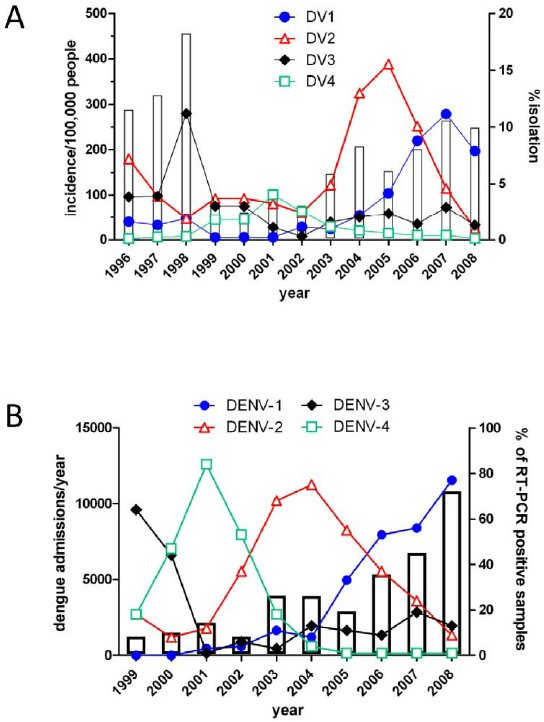
Sequential infection increases the risk of a severe form of the infection - dengue hemorrhagic fever.
The dengue virus
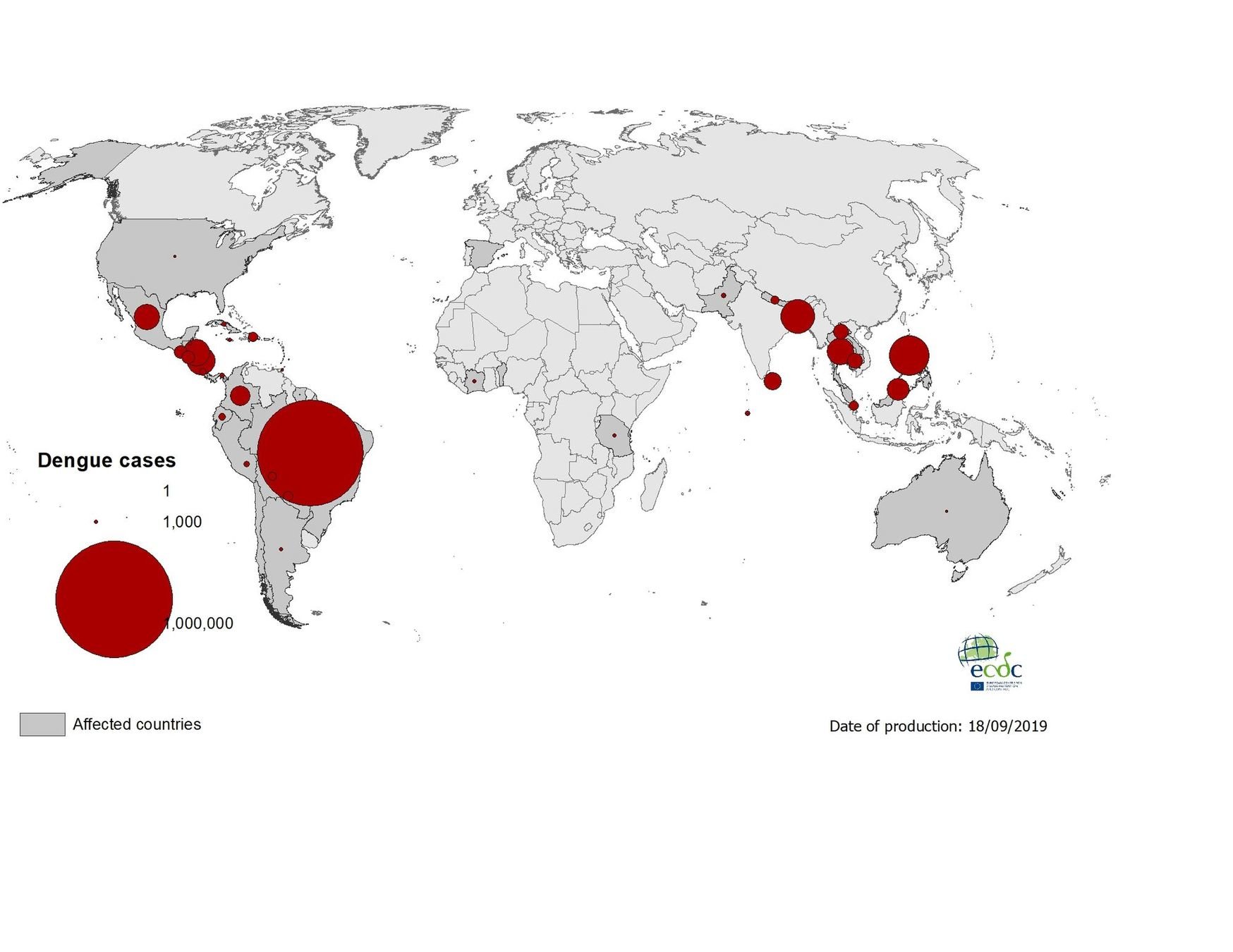

Geographical distribution of dengue cases reported worldwide, July to September 2019
17 September 2019 - laboratory-confirmed autochthonous case of dengue in Barcelona, Spain
| Transmission & Disease
The dengue virus

Geographical distribution of dengue cases reported worldwide, January to February 2020
| Transmission & Disease
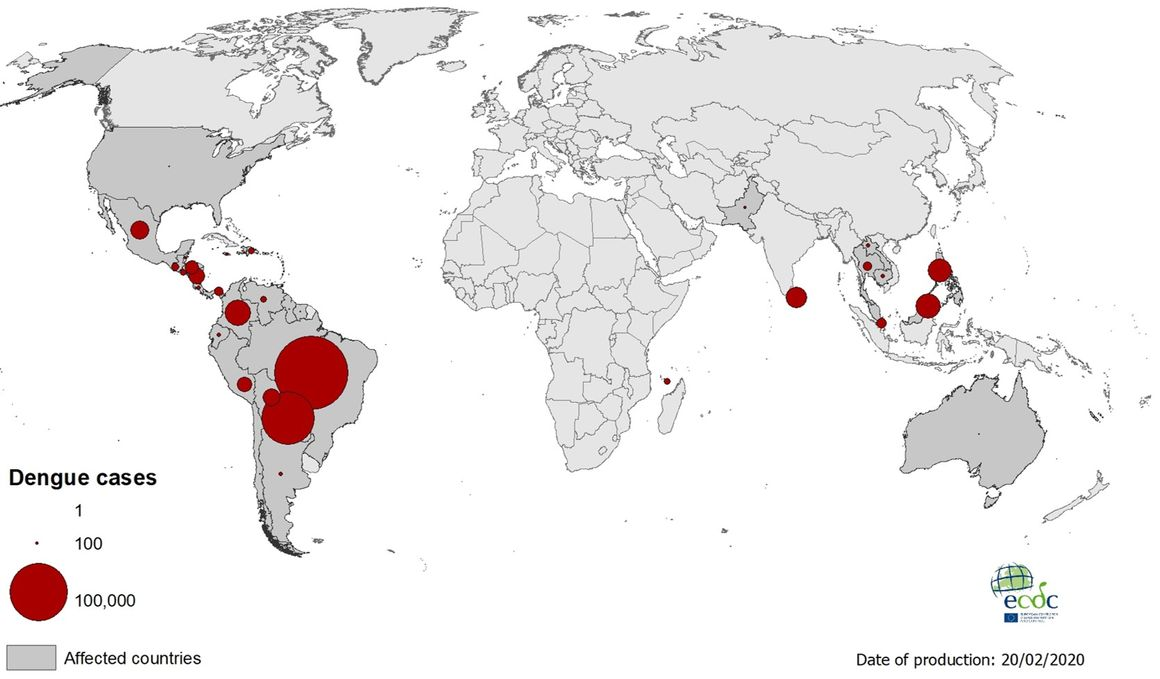
The four dengue virus serotypes are currently circulating simultaneously in the Region of the Americas which increases the risk of severe cases.
The dengue virus
| The virus
Dengue hemorrhagic fever:
- The virus presents 4 different serotypes, sub-classified in many genotypes
- After "primary" infection with one serotype, "secondary" infections by one or more of the other serotypes can precipitate ‘antibody dependant enhancement’ (ADE)
- Infection with one type gives little immune protection against the other types
The dengue virus
| The virus
- DENV-1 - genotypes I-V
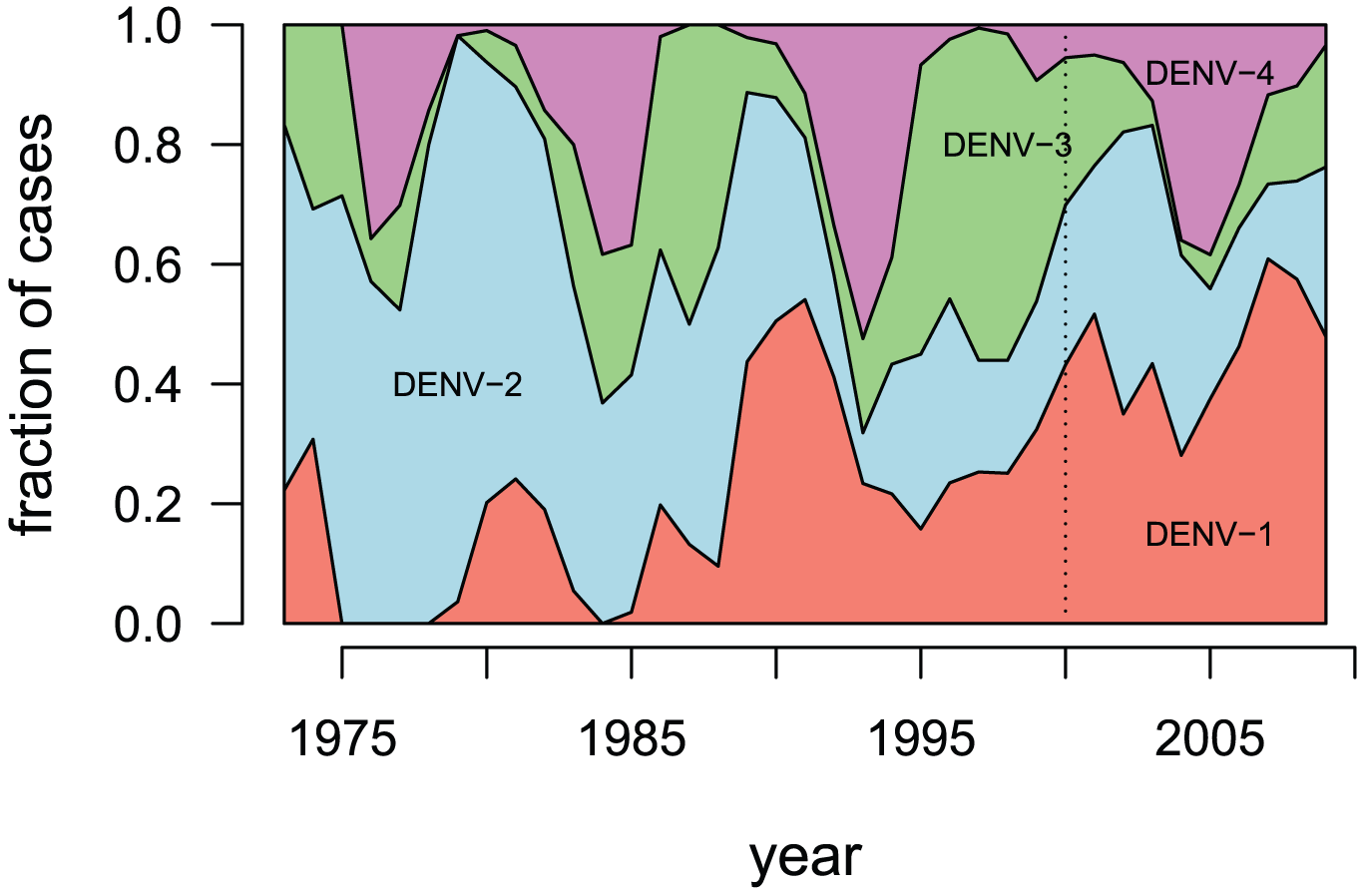
- DENV-2 - genotypes Asian I, Asian II, Cosmopolitan, American, Asian/American & Sylvatic
- DENV-3 - genotypes I-V
- DENV-4 - genotypes I - III & Sylvatic
https://doi.org/10.1371/journal.pntd.0001876.g002
https://doi.org/10.1371/journal.pntd.0000757
Thailand
Viet Nam
The dengue virus
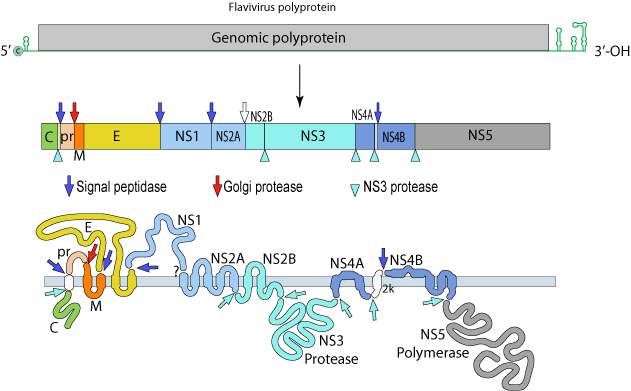
DENV: (+)ssRNA (~11Kb; 1 ORF)
The single polyprotein encodes:
-
Structural Proteins:
-
C – capsid
-
prM – pre-membrane
-
M - membrane
-
E - envelope
-
-
Non-Structural Proteins:
-
NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5
-

| The virus
The dengue virus
| The virus
Empower the use of HTS to monitor the dissemination of the disease
RNA Extraction
PCR
Amplification
HTS Sequencing
➔
➔
How do the different genotypes model transmission and infection?

| The Workflow
- Ready to use but customizable
- Scalable
- Reproducible
- Stand-alone (confidential data)
- Easy to explore and share results
Requirements


A solution





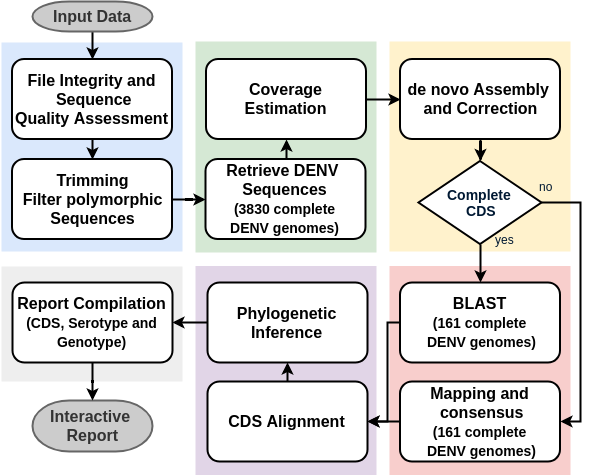
A ready-to-use, reproducible, scalable workflow for the identification, serotyping, genotyping and phylogenetic analysis of DENV from HTS data
| The Workflow




DENV Identification
- 3830 complete DENV genomes from the NIAID Virus Pathogen Database and Analysis Resource (ViPR)
- complete genome sequence
- human host (exception of DENV-1 III, monkey)
- collection year (1950-2018)
In Silico Typing:
- 161 representative sequences of all sero and genotypes.
| The Workflow

- DENV-1


- DENV-2
a) Envelope Region b) whole genome sequence
| The Workflow

- DENV-3


- DENV-4
a) Envelope Region b) whole genome sequence
| The Workflow

| Test datasets

Shotgun Metagenomics dataset:
- 9 plasma samples
- 13 serum samples
- 1 spiked sample with the 4 serotypes
- Positive and Negative controls


nextflow run DEN-IM.nf -profile slurm_shifter --fastq="fastq/*_{1,2}.*"- HPC with 300 Cores/Processing Power and 3 TB RAM
- On average, each sample took 7 minutes to analyse. A total of 75 CPU hours were used to analyse the 25 samples, with a total of 17Gb in size. This analysis resulted in 69Gb of data.


| Test datasets
| Where to find it?




Git, Nextflow (java) and a container engine (Docker, singularity, shifter...).
apt-get install gitcurl -s https://get.nextflow.io | bash
apt-install docker-ce
Clone (or run remotely)
git clone https://github.com/B-UMMI/DEN-IM.git| Where to find it?




https://github.com/B-UMMI/DEN-IMhttps://github.com/B-UMMI/DEN-IM/wiki
This work was funded by: FCT - "Fundação para a Ciência e a Tecnologia" (SFRH/BD/129483/2017) and the Abel Tasman Talent Program grant from the UMCG, University of Groningen, Groningen, The Netherland
Special thanks to E Lizarazo, M P Machado, D N Silva, A Tami, M Ramirez, N Couto, J W A Rossen, J A Carriço






