
Inês Mendes
MRamirez Lab - iMM


@ines_cim
cimendes
XIV CAML · IV NeurULisboa PhD Students Meeting
5th to 7th May, 2021
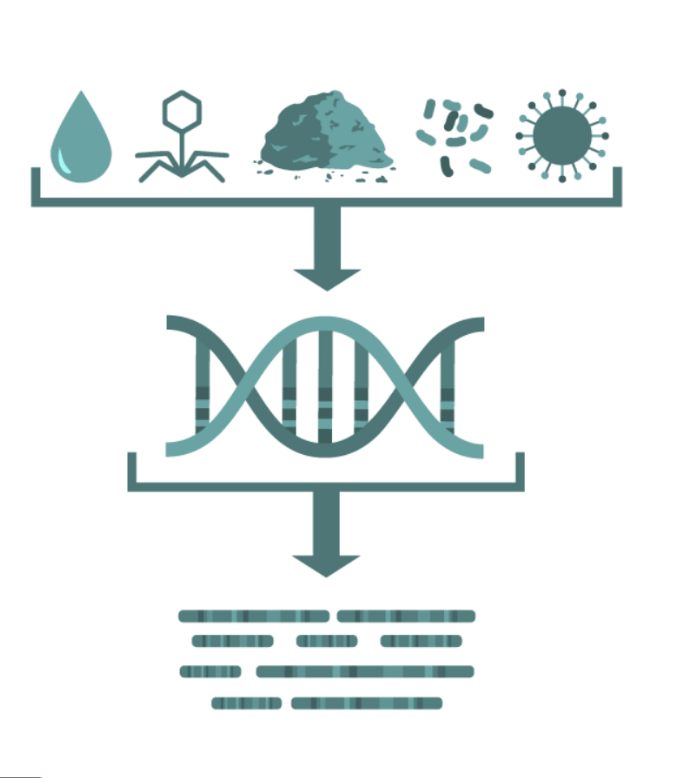
Metagenomics
Random "shotgun" sequencing of microbial DNA, without selecting a particular gene or species.
Promising methodology for obtaining fast results for the identification of pathogens and their virulence and antimicrobial resistance properties without the need for culture.
| The motivation
Metagenomics
| The motivation
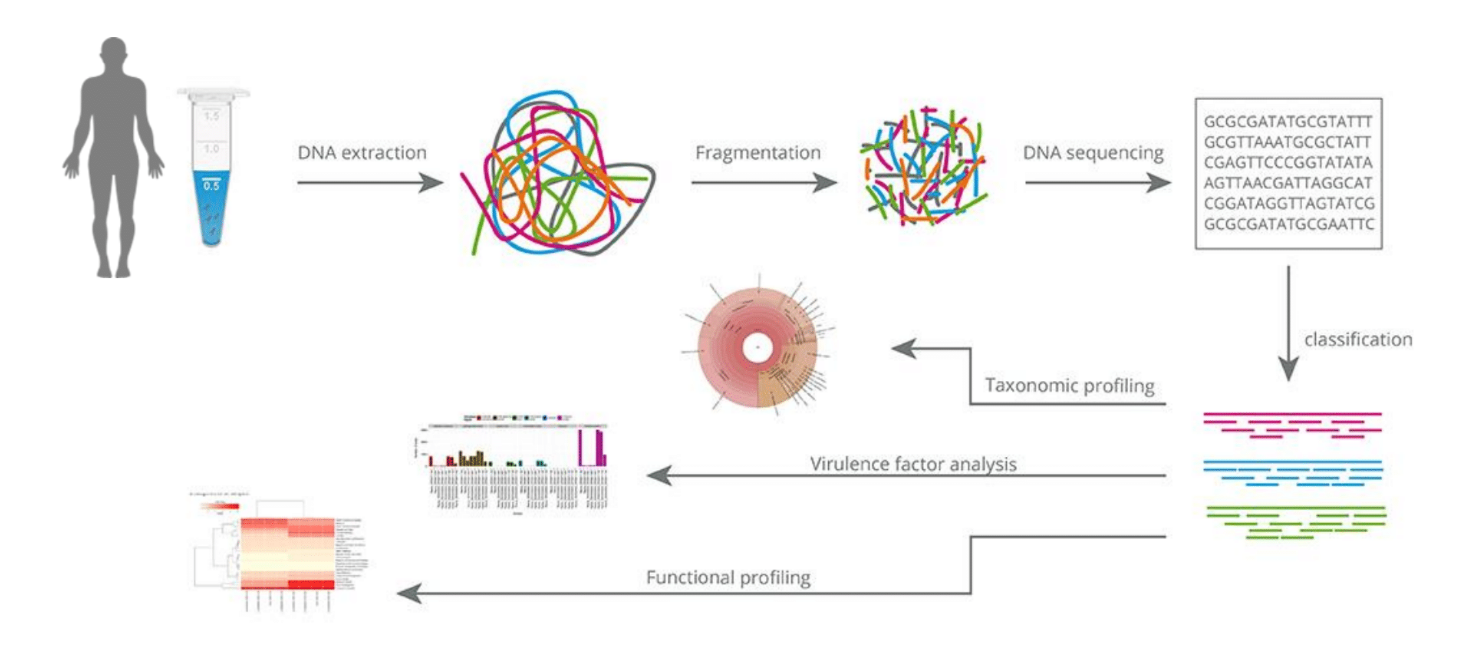

de novo Assembly
The assembly methods provide longer sequences that are more informative than shorter sequencing data and can provide a more complete picture of the microbial community in a given sample.
Metagenomics
| An assembly challenge
- Results are highly dependent on the tools chosen for the analysis - Lack of standardization and proper benchmark.
Major issues
Reads
Contigs
Genomes
- Highlights the potential and the limitations of shotgun metagenomics as a diagnostic tool - Lack of reproducibility
LMAS
| de novo Assembly Benchmark
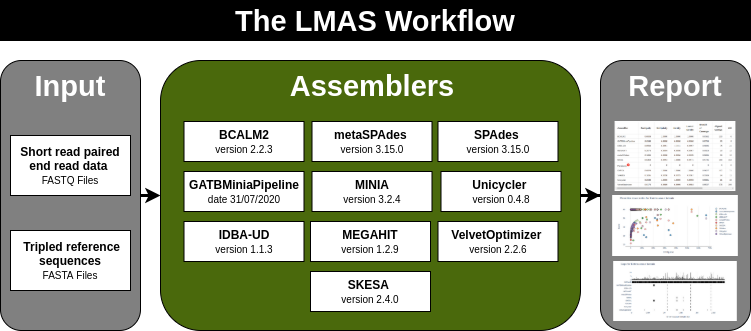

https://github.com/cimendes/LMAS

https://lmas.readthedocs.io/
LMAS
| de novo Assembly Benchmark






Git, Nextflow (java) and a container engine (Docker, singularity, shifter...).
apt-get install gitcurl -s https://get.nextflow.io | bash
apt-install docker-ceClone
git clone https://github.com/cimendes/LMAS.gitRun LMAS
nextflow run LMAS.nfLMAS
| de novo Assembly Benchmark
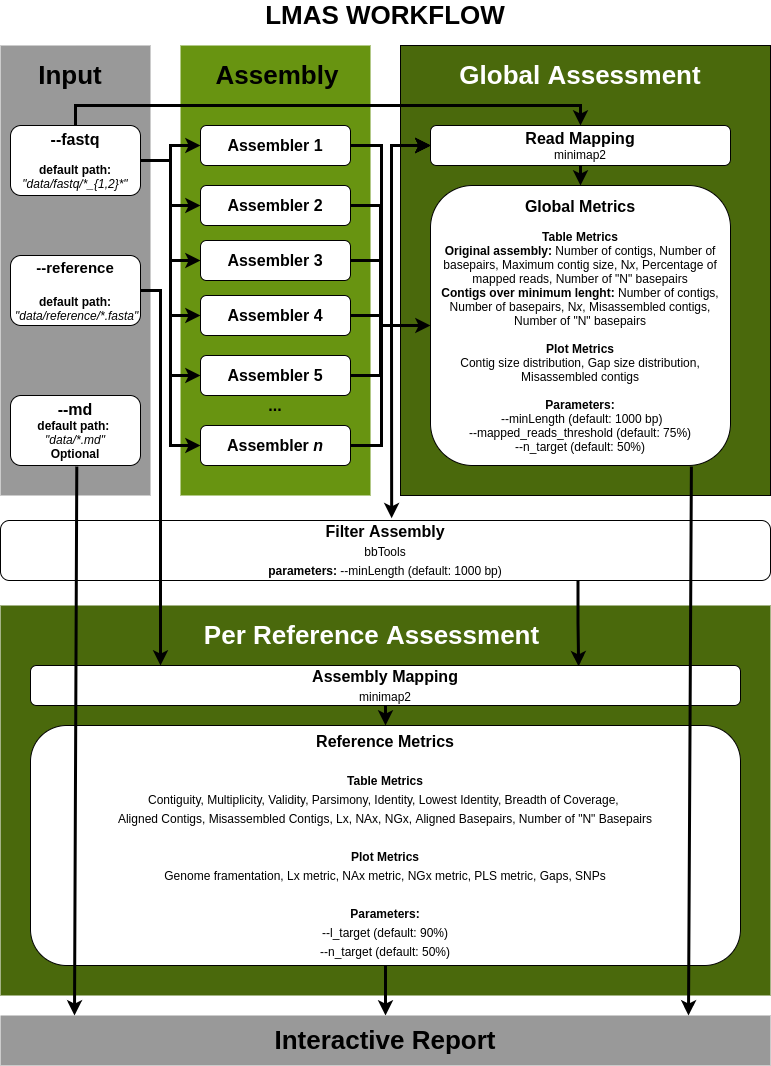
The input data is assembled in parallel by the set of genomic and metagenomic de novo assemblers in LMAS.
The global and per reference metrics are grouped in the interactive LMAS report for exploration
The resulting assembled sequences are processed and assembly quality metrics are computed,
LMAS
| ZymoBIOMICS Standards

The resulting LMAS report is available at
Eight bacterial genomes and four plasmids of the ZymoBIOMICS Microbial Community Standards were used as the triple reference.
| Sample | Distribution | Error Model | Read Pairs (M) |
|---|---|---|---|
| ENN | Even | None | 8.6 |
| EHS | Even | Illumina HiSeq | 8.6 |
| ERR2984773 | Even | Real Sample | 8.6 |
| LNN | Log | None | 47.5 |
| LHS | Log | Illumina HiSeq | 47.5 |
| ERR2935805 | Log | Real Sample | 47.5 |
Special thanks to Pedro Vila-Cerqueira, Rafael Maria Mamede and Mário Ramirez.







Thank you for your attention


FCT PhD Grant SFRH/BD/129483/2017