Reproducibility in Computational Biology
Medical Microbiology Maastricht UMC
May 26th, 2020


@ines_cim
cimendes
Inês Mendes




@ines_cim
cimendes

Who am I
| Casper made me do this
- BSc in Cellular and Molecular Biology (New University of Lisbon)
- MSc in Bioinformatics and Computational Biology (University of Lisbon)
- 3rd year PhD slave in Bioinformatics for Clinical Microbiology (University of Lisbon; University of Groningen)
- Dog Lover ♥


Reproducibility
| The questions
Reproducibility
| The questions
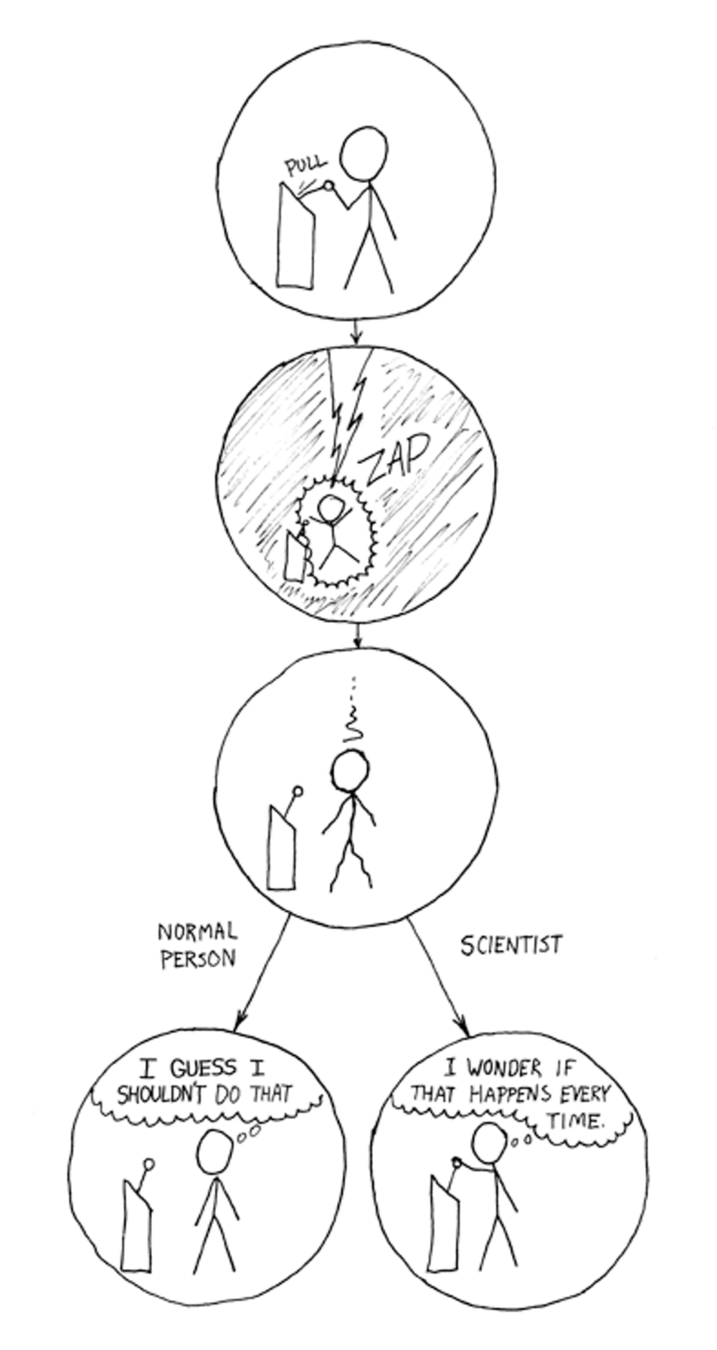
Can person X, with the same data and the same methodology, obtain the same conclusions?
Reproducibility
| The questions


- Do I know what is happening?
- Is it reproducible?
- Is it shareable?


Black
Box
Transparent
Box
- Commercial/Freeware
- You get what it gives you
- Ready to use
- Stealth change
- Standalone
- Freeware
- You can "tailor"
- "Major" headache
- Visible change
- Dependencies
The needs:
- Analyze a large amount of sequence data routinely
- Some computationally intensive steps
- Constantly updating/adding software
Reproducibility
| The needs
Writing of pipelines in python/perl/shell scripts circa 2000, colorized.

- Custom ad-hoc scripts
- Difficult to parallelise
- Difficult to install/run
- Hard to deploy in multiple environments
- What's workflow managers?!
- What's docker?!
Reproducibility
| The needs
Workflows in the Paleolithic era:
The game changing combination of workflow managers and containers:
- Portability
- Reproducible
- Scalability
- Multi-scale containerization
- Native cloud support
Reproducibility
| The needs
Workflows in the Modern era:







Reproducibility
| The challenges
- No standard way of describing experiments, environments, (derived) data, and workflows.
- No transparency in creating environments and steps/methods to recreate analysis.
- The experimental nature of the research code and ecosystem makes it often hard to build.
- Unresolved or undocumented dependencies.
- Infrastructure for storage and distribution.
Reproducibility
| The challenges
- No standard way of describing experiments, environments, (derived) data, and workflows.
- No transparency in creating environments and steps/methods to recreate analysis.
- The experimental nature of the research code and ecosystem makes it often hard to build.
- Unresolved or undocumented dependencies.
- Infrastructure for storage and distribution.
What is our role, as computational biologists, to change this?
Reproducibility
| The ideal scenario
Version Control
| What is it?




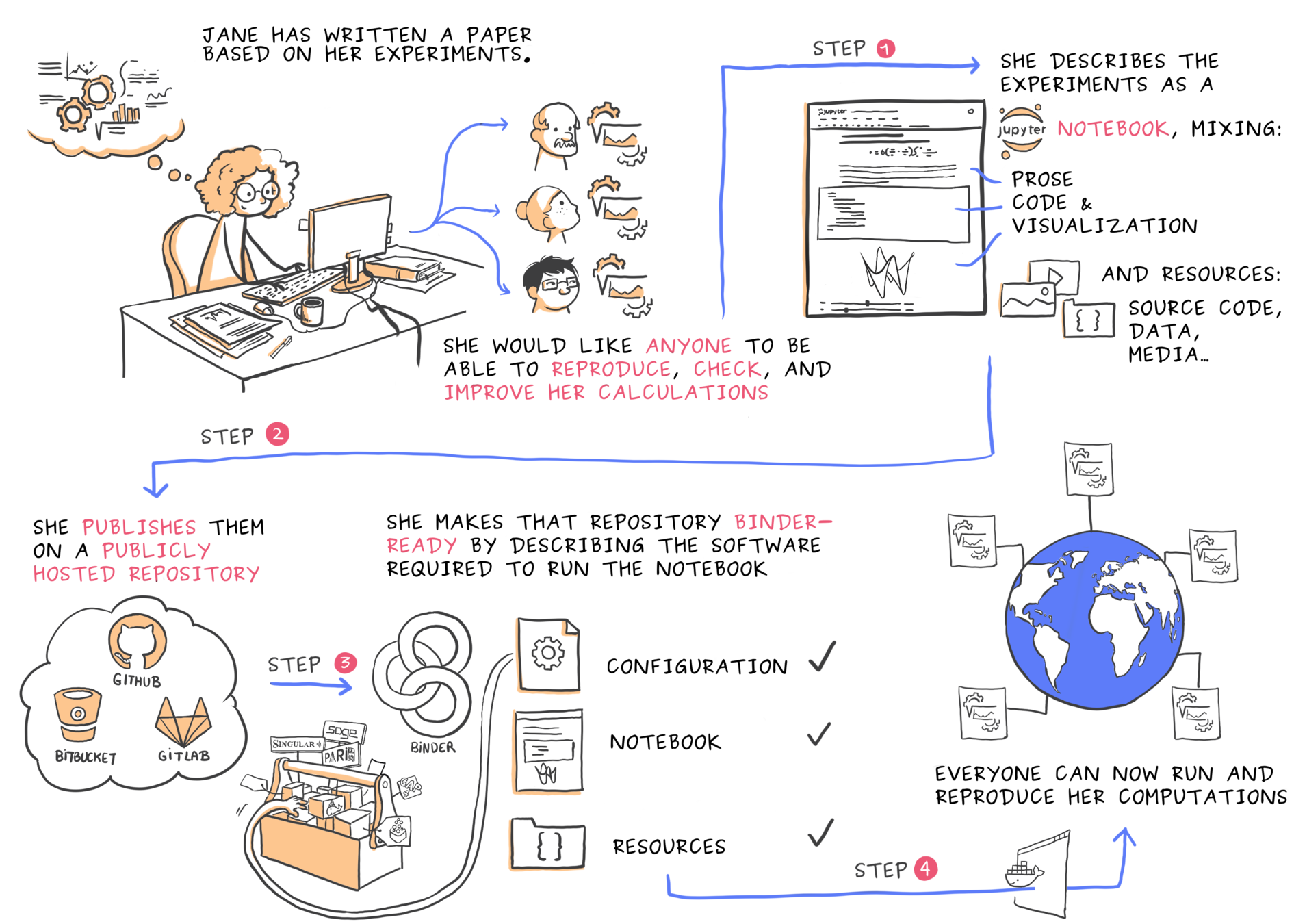
It records changes to a file or set of files over time so that you can recall specific versions later.
It allows you to revert selected files back to a previous state, revert the entire project back to a previous state, compare changes over time, see who last modified something that might be causing a problem, who introduced an issue and when, and more.
Version Control
| What is it?
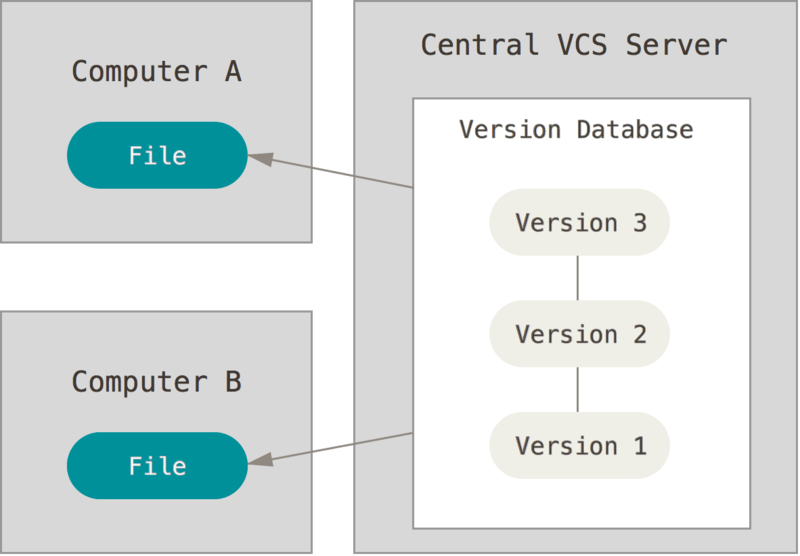






Version Control
| What is it?
Version Control
| What is it?
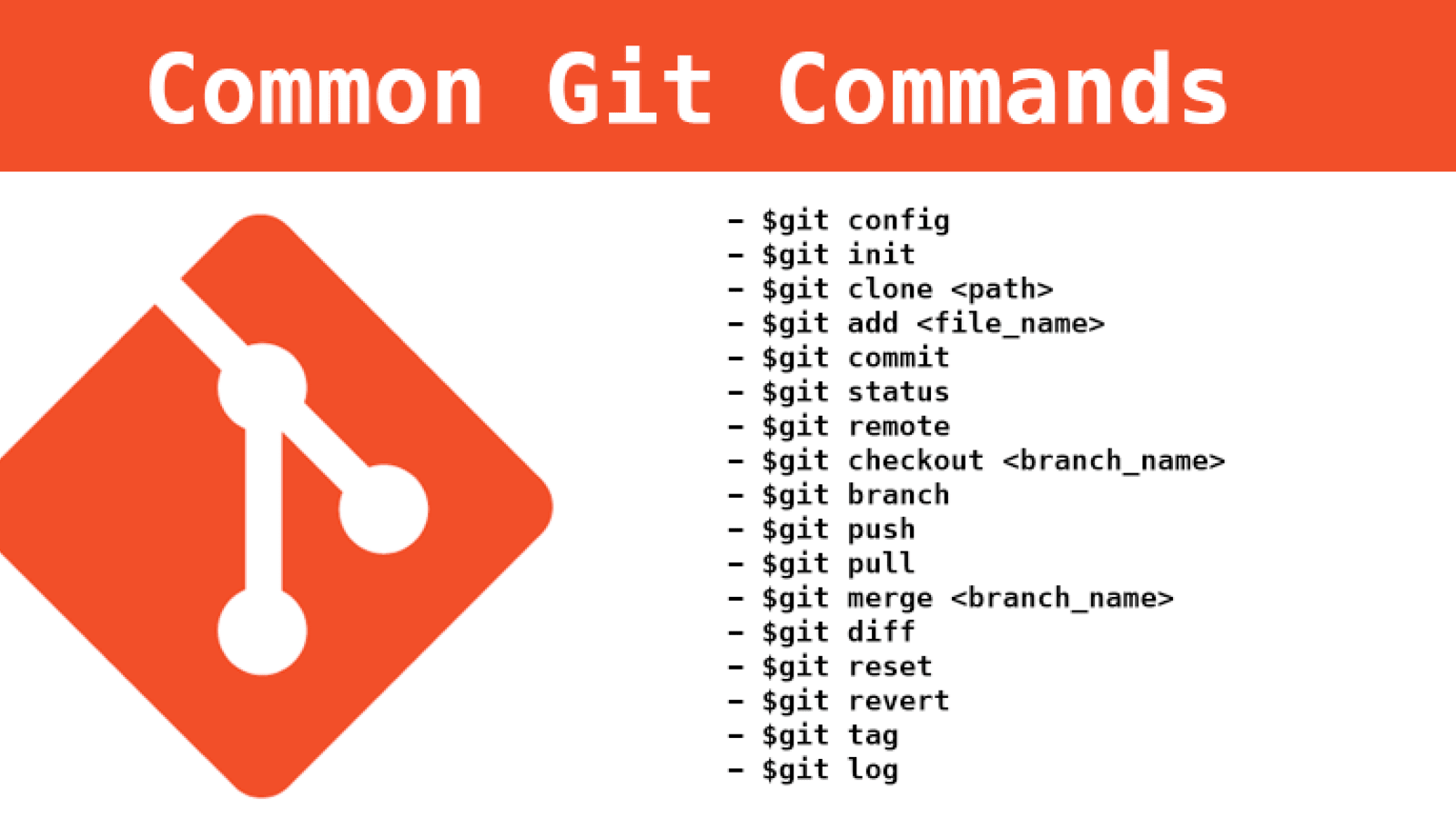
Software Containers


Runs the same regardless of the environment.
Enables the distribution and deployment of scientific software in a runnable state.
A container image is a lightweight, stand-alone, executable package of a software that includes everything needed to run it:
- code
- runtime
- system tools
- system libraries
| What is it?
Host Hardware
Host Hardware
Container Engine
Host OS
Host OS
Hypervisor
Guest OS
Guest OS
App
App
Guest OS
VM1
VM2
App
App
App
App
Virtual Machines
Containers
Software Containers
| VMs and Containers
Software Containers
| Solutions
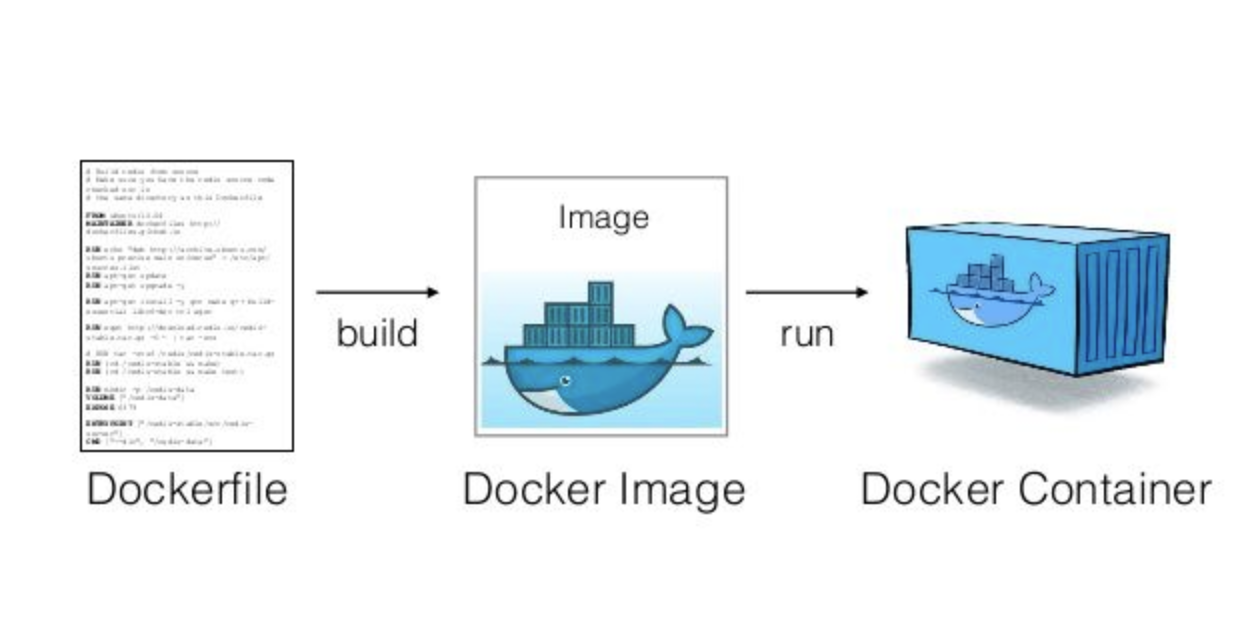


Docker Hub
build
pull & run
host
push
Workflow Managers
| What is it?



Enables scalable and reproducible scientific workflows. It simplifies the deployment of complex parallel and reactive workflows.
Reactive workflow framework
Create pipelines with asynchronous (and implicitly parallelized) data streams
Programing DSL
Has its own language for building a pipeline
Containerized
Out of the box integration with containers engines (Docker, Singularity, Shifter)
The creation of workflow pipelines was designed for bioinformaticians familiar with programming.
It's execution is for everyone.
Workflow Managers
| What is it?
- Automatic management of temporary input/output directory/files
- No need for custom handling of concurrency (parallelization)
- A single pipeline can support any scripting language (Bash, Python, Perl, R...)
- Every process (task) can be run in a container
- It's portability allows for the same pipeline to run on a laptop, server, cluster, etc
- Checkpoints and resume functionality
- Host pipeline on GitHub and run remotely




Monolithic pipelines


Reproducibility
| The happy mariage
Need to change often
Siloed tool containers
Don't do much by themselves
Use cases
|
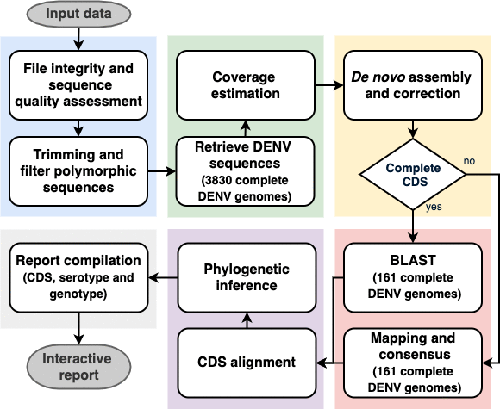





https://github.com/B-UMMI/DEN-IM.gitUse cases
| Innuendo Platform

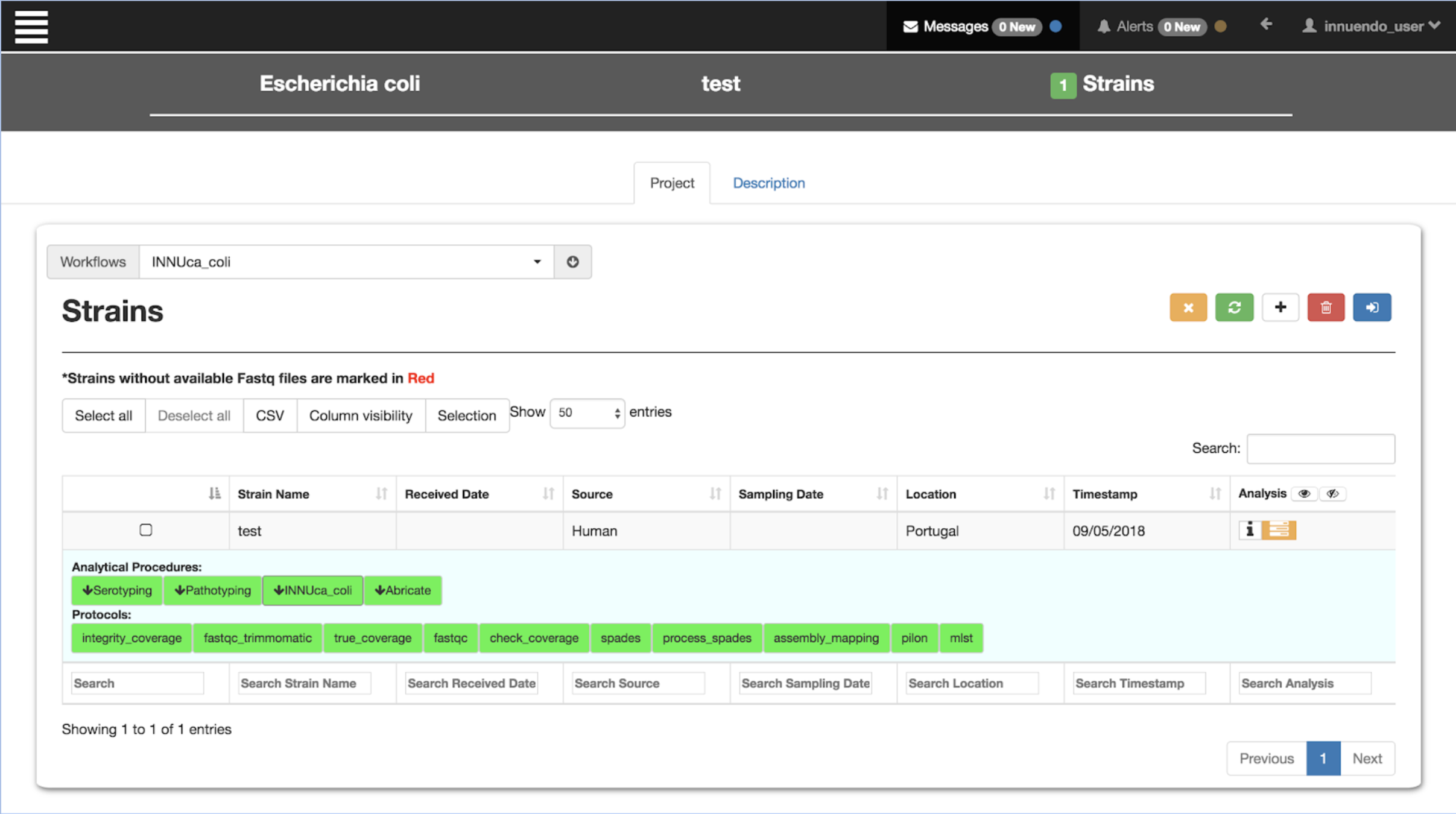




Use cases
|

Workflow based development
Component based development
Components are modular pieces with some basic rules:

Component A
- Input/Output
- Parameters
- Resources
Component B

- Input/Output
- Parameters
- Resources



Use cases
|

With this framework, building workflows becomes simple:
flowcraft build -t 'trimmomatic fastqc spades pilon' -o my_nextflow_pipeline
Results in the following workflow DAG (direct acyclic graph)
It's easy to get experimental:
flowcraft build -t 'trimmomatic fastqc skesa pilon' -o my_nextflow_pipeline
Switch spades for skesa



Use cases
| Genomics & Epidemiology

42h on 200 CPUs
151 samples
1812 assemblies
43s/assembly
Sampled assemblies


Use cases
| Genomics & Epidemiology


Dots above red line
Same sample interpreted with different profile
Potentially undetected outbreak

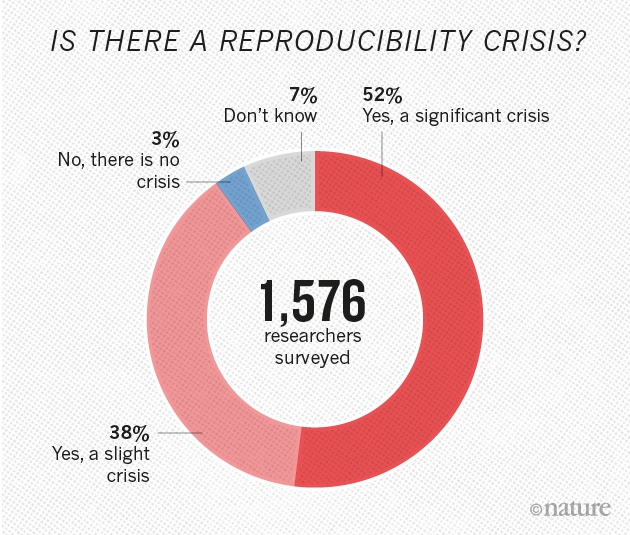
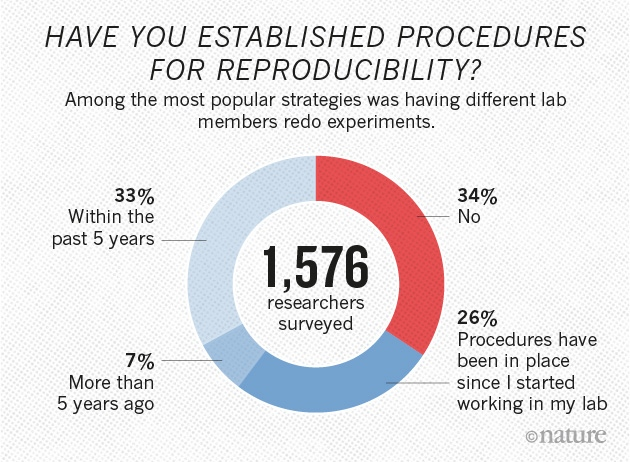
This work was funded by: FCT - "Fundação para a Ciência e a Tecnologia" (SFRH/BD/129483/2017)
Special thanks to Diogo Silva, Bruno Gonçalves, Tiago Jesus, Pedro Vila-Cerqueira, Rafael Maria Mamede, João Carriço, John Rossen and Mário Ramirez.






