Workflow automation with
Nextflow and Flowcraft


3rd Meeting Bioinformatics in Medical Microbiology NL
@ines_cim
cimendes
Inês Mendes




- Do I know what is happening?
- Is it reproducible?
- Is it shareable?

Reproducibility
| The questions

Black
Box
Transparent
Box
- Commercial/Freeware
- You get what it gives you
- Ready to use
- Stealth change
- Standalone
- Freeware
- You can "tailor"
- "Major" headache
- Visible change
- Dependencies
The needs:
- Analyze a large amount of sequence data routinely
- Some computationally intensive steps
- Constantly updating/adding software
Reproducibility
| The needs
Reproducibility
| The challenges
- No standard way of describing experiments, environments, (derived) data, and workflows.
- No transparency in creating environments and steps/methods to recreate analysis.
- The experimental nature of the research code and ecosystem makes it often hard to build.
- Unresolved or undocumented dependencies.
- Infrastructure for storage and distribution.
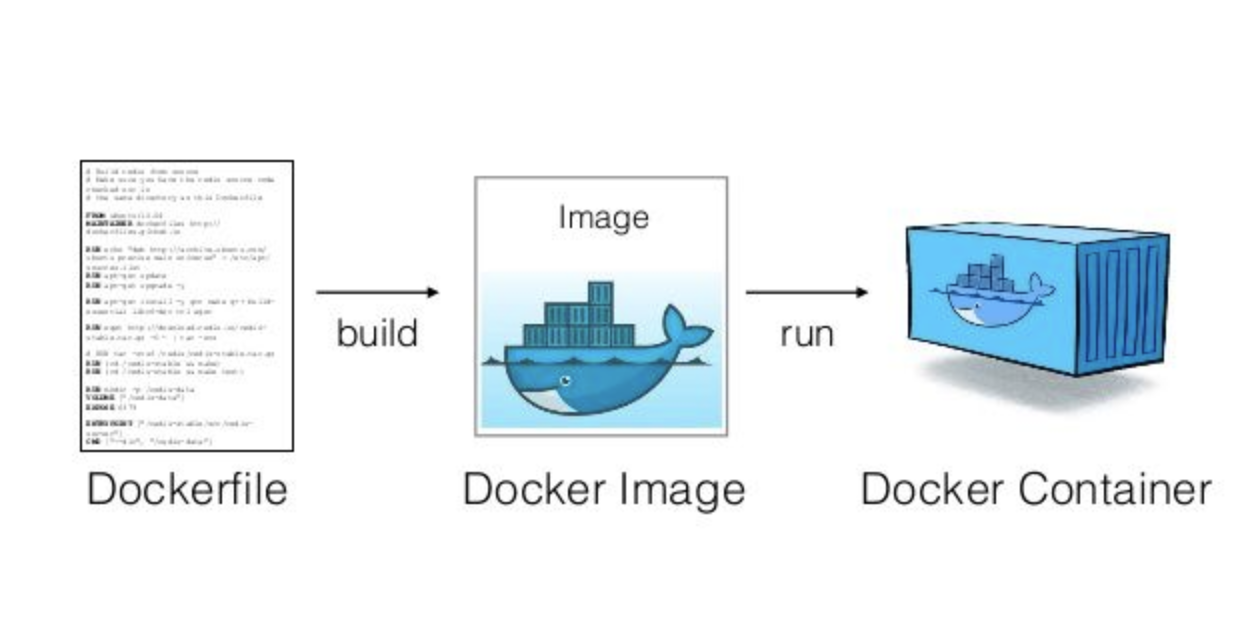
Software Containers


Runs the same regardless of the environment.
Enables the distribution and deployment of scientific software in a runnable state.
A container image is a lightweight, stand-alone, executable package of a software that includes everything needed to run it:
- code
- runtime
- system tools
- system libraries
| What is it?
Host Hardware
Host Hardware
Container Engine
Host OS
Host OS
Hypervisor
Guest OS
Guest OS
App
App
Guest OS
VM1
VM2
App
App
App
App
Virtual Machines
Containers
Software Containers
| VMs and Containers
Software Containers
| Solutions


Docker Hub
build
pull & run
host
push
Nextflow
| What is it?
Enables scalable and reproducible scientific workflows using software containers. It simplifies the deployment of complex parallel and reactive workflows.
Reactive workflow framework
Create pipelines with asynchronous (and implicitly parallelized) data streams
Programing DSL
Has its own language for building a pipeline
Containerized
Out of the box integration with containers engines (Docker, Singularity, Shifter)

Nextflow
| Requirements
Installation
- Bash
Ubiquitous on UNIX. Windows: Cygwin or Linux sysbsystem, maybe...- Java 1.8
sudo apt-get install openjdk-8-jdk- Nextflow
curl -s https://get.nextflow.io | bashOptional (but recommended)
- Container engine (Docker, Singularity...)
The creation of Nextflow pipelines was designed for bioinformaticians familiar with programming.
It's execution is for everyone.

Nextflow
| Why bother?
- Automatic management of temporary input/output directory/files
- No need for custom handling of concurrency (parallelization)
- A single pipeline can support any scripting language (Bash, Python, Perl, R...)
- Every process (task) can be run in a container
- It's portability allows for the same pipeline to run on a laptop, server, cluster, etc
- Checkpoints and resume functionality
- Steaming capability
- Host pipeline on GitHub and run remotely

Creating a pipeline
| Key concepts
Processes
- Building blocks of the pipeline - Can contain Unix shell commands or code written in any scripting language (Python, Perl, R, Bash, etc).
- Executed independently and in parallel.
- Contain directives for container to use, CPU/RAM/Disk limits, etc
Process A
Process B
Creating a pipeline
| Key concepts
Processes
- Building blocks of the pipeline - Can contain Unix shell commands or code written in any scripting language (Python, Perl, R, Bash, etc).
- Executed independently and in parallel.
- Contain directives for container to use, CPU/RAM/Disk limits, etc
Process A
Process B
Channels
- Unidirectional FIFO communication channels between processes
Channel 1
Channel 2
Input
Creating a pipeline
| Process
process <name>{
input:
<input channels>
// Optional
output:
<output channels>
script:
"""
<command/code block>
"""
}1. Set process name
2. Set one or more input channels
3. Output channels are optional
4. The code block is always last
Creating a pipeline
| Process
process <name>{
input:
<input channels>
// Optional
output:
<output channels>
script:
"""
template my_script.py
"""
}1. Set process name
2. Set one or more input channels
3. Output channels are optional
4. Or use a template
#!/usr/bin/python
# Store in templates/my_script.py
def main():
print("Hello world!")
main()Creating a pipeline
| Process
process anExample{
input:
val x from Channel.value(1)
file y from Channel.fromPath("/path/to/file")
// Optional
output:
val "some_str" into outputChannel
script:
"""
template my_script.py
"""
}#!/usr/bin/python
# Store in templates/my_script.py
def main():
print("The input variables are ${x} and ${y}")
main()Creating a pipeline
| An example
startChannel = Channel.fromFilePairs(params.fastq)
process fastQC {
input:
set sampleId, file(fastq) from startChannel
output:
set sampleId, file(fastq) into fastqcOut
"""
fastqc --extract --nogroup --format fastq \
--threads ${task.cpus} ${fastq}
"""
}
process mapping {
input:
set sampleId, file(fastq) from fastqcOut
each file(ref) from Channel.fromPath(params.reference)
"""
bowtie2-build --threads ${task.cpus} \
${ref} genome_index
bowtie2 --threads ${task.cpus} -x genome_index \
-1 ${fastq[0]} -2 ${fastq[1]} -S mapping.sam
"""
}FastQC
mapping
startChannel
fastqcOutput
reference
Creating a pipeline
| Customize it
process mapping {
container "ummidock/bowtie2_samtools:1.0.0-1"
cpus 1
memory "1GB"
publishDir "results/bowtie/"
input:
set sampleId, file(fastq) from fastqcOut
each file(ref) from Channel.fromPath(params.reference)
output:
file '*_mapping.sam'
"""
<command block>
"""
}FastQC
mapping
- Set different containers for each process
- Set custom CPU/RAM profiles that maximize pipeline performance
- More options at directives documentation
CPU: 2
Mem: 3GB
CPU: 1
Mem: 1GB
Creating a pipeline
| The config file
// nextflow.config
params {
fastq = "data/*_{1,2}.*"
reference = "ref/*.fasta"
}
process {
$fastQC.container = "ummidock/fastqc:0.11.5-1"
$mapping.container = "ummidock/bowtie2_samtools:1.0.0-1"
$mapping.publishDir = "results/bowtie/"
$fastQC.cpus = 2
$fastQC.memory = "2GB"
$mapping.cpus = 4
$mapping.memory = "2GB"
}
profiles {
standard {
docker.enabled = true
}
}nextflow.config
- Parameters
- Process directives
- Docker options
- Profiles
Creating a pipeline
| Channel operators
fastQC
startChannel
fastqcOut
mapping
Operatos can transform and connect other channels without interfering with processes
There are dozen of available operators
Creating a pipeline
| Channel operators
Channel
.from( 'a', 'b', 'aa', 'bc', 3)
.filter( ~/^a.*/ )
.into { newChannel }
a
aa
Channel
.from( 'a', 'b', 'aa', 'bc', 3)
.into { newChannel1; NewChannel2 }
Channel
.from( 1, 2, 3, 4 )
.collect()
.into { OneChannel }
[1,2,3,4]Channel
.from( 1, 2, 3 )
.map { it * it }
.into {newChannel}
1
4
9
Filter channels
Map channels
Fork channels
Collect channels
More on Nextflow
- Nextflow website:
- https://www.nextflow.io/
- Nextflow docs:
- https://www.nextflow.io/docs/latest/index.html
- Awesome nextflow:
- https://github.com/nextflow-io/awesome-nextflow
FlowCraft
| The motivation
The game changing combination of nextflow + containers:
- Portability
- Reproducible
- Scalability
- Multi-scale containerization
- Native cloud support




Substantial challenges still persist:
- Fast pace of bioinformatics software landscape
- Continuous need for benchmarking and comparative analyses
- The need for agile and dynamic pipeline building
- Remove the pain of changing inner workings of workflows
FlowCraft
| The premise
Workflow based development
Component based development
Components are modular pieces of nextflow code with some basic rules:

Component A
- Input/Output
- Parameters
- Resources
Component B

- Input/Output
- Parameters
- Resources
FlowCraft
| The premise
With this framework, building workflows becomes simple:
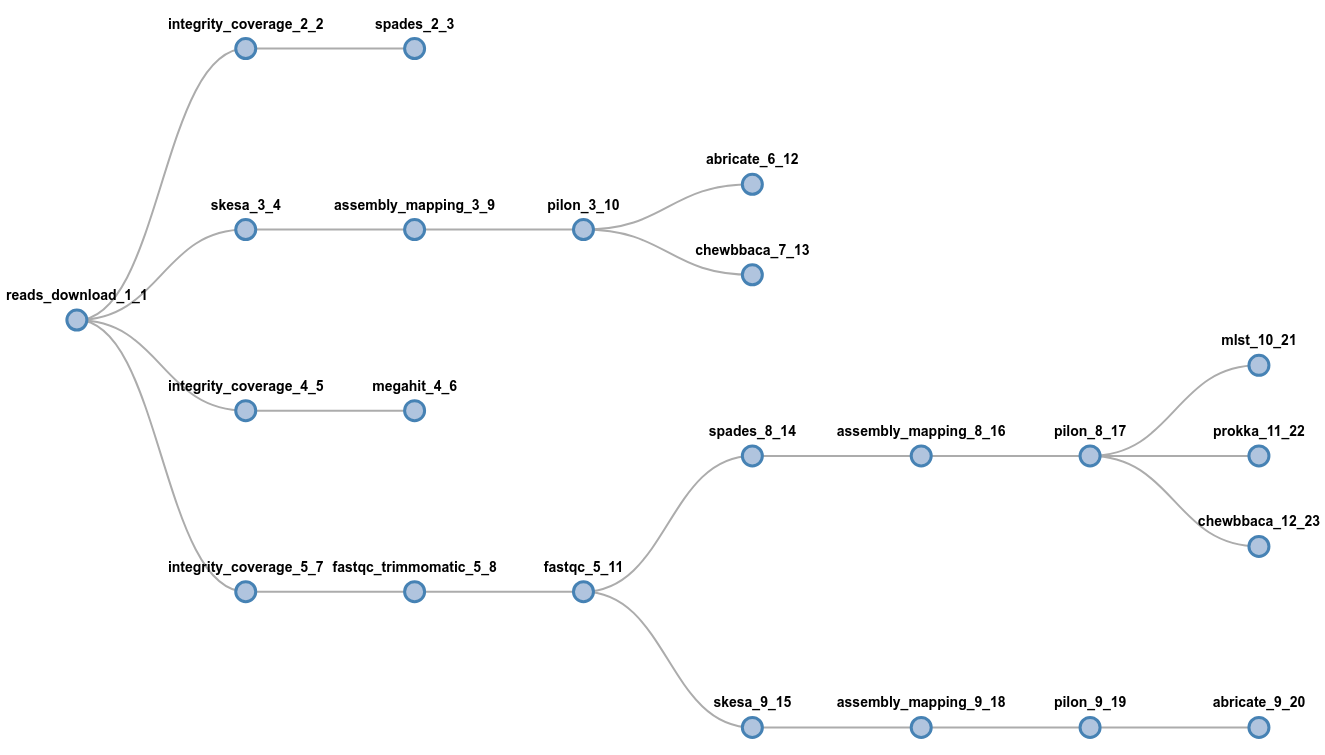
flowcraft build -t 'trimmomatic fastqc spades pilon' -o my_nextflow_pipeline
Results in the following workflow DAG
$ nextflow run my_nextflow_pipeline.nf --help
N E X T F L O W ~ version 0.32.0
Launching `my_nextflow_pipeline.nf` [jovial_swirles] - revision: b4473f5a12
============================================================
F L O W C R A F T
============================================================
Built using flowcraft v1.4.0
Usage:
nextflow run my_nextflow_pipeline.nf
--fastq Path expression to paired-end fastq files. (default: fastq/*_{1,2}.*) (default: 'fastq/*_{1,2}.*')
Component 'INTEGRITY_COVERAGE_1_1'
----------------------------------
--genomeSize_1_1 Genome size estimate for the samples in Mb. It is used to estimate the coverage and other assembly parameters andchecks (default: 1)
--minCoverage_1_1 Minimum coverage for a sample to proceed. By default it's setto 0 to allow any coverage (default: 0)
Component 'TRIMMOMATIC_1_2'
---------------------------
--adapters_1_2 Path to adapters files, if any. (default: 'None')
--trimSlidingWindow_1_2 Perform sliding window trimming, cutting once the average quality within the window falls below a threshold (default: '5:20')
--trimLeading_1_2 Cut bases off the start of a read, if below a threshold quality (default: 3)
--trimTrailing_1_2 Cut bases of the end of a read, if below a threshold quality (default: 3)
--trimMinLength_1_2 Drop the read if it is below a specified length (default: 55)
--clearInput_1_2 Permanently removes temporary input files. This option is only useful to remove temporary files in large workflows and prevents nextflow's resume functionality. Use with caution. (default: false)
Component 'FASTQC_1_3'
----------------------
--adapters_1_3 Path to adapters files, if any. (default: 'None')
Component 'SPADES_1_4'
----------------------
--spadesMinCoverage_1_4 The minimum number of reads to consider an edge in the de Bruijn graph during the assembly (default: 2)
--spadesMinKmerCoverage_1_4 Minimum contigs K-mer coverage. After assembly only keep contigs with reported k-mer coverage equal or above this value (default: 2)
--spadesKmers_1_4 If 'auto' the SPAdes k-mer lengths will be determined from the maximum read length of each assembly. If 'default', SPAdes will use the default k-mer lengths. (default: 'auto')
--clearInput_1_4 Permanently removes temporary input files. This option is only useful to remove temporary files in large workflows and prevents nextflow's resume functionality. Use with caution. (default: false)
--disableRR_1_4 disables repeat resolution stage of assembling. (default: false)
Component 'ASSEMBLY_MAPPING_1_5'
--------------------------------
--minAssemblyCoverage_1_5 In auto, the default minimum coverage for each assembled contig is 1/3 of the assembly mean coverage or 10x, if the mean coverage is below 10x (default: 'auto')
--AMaxContigs_1_5 A warning is issued if the number of contigs is overthis threshold. (default: 100)
--genomeSize_1_5 Genome size estimate for the samples. It is used to check the ratio of contig number per genome MB (default: 2.1)
Component 'PILON_1_6'
---------------------
--clearInput_1_6 Permanently removes temporary input files. This option is only useful to remove temporary files in large workflows and prevents nextflow's resume functionality. Use with caution. (default: false)Help and parameters tailor-made to the pipeline
FlowCraft
| The premise
It's easy to get experimental:
flowcraft build -t 'trimmomatic fastqc skesa pilon' -o my_nextflow_pipeline
Switch spades for skesa
flowcraft build -t 'trimmomatic fastqc skesa pilon (abricate | prokka)' -o my_nextflow_pipelineAdd genome annotation components in the end

FlowCraft
| The premise
It's easy to get wild:
flowcraft build -t 'reads_download (
spades | skesa pilon (abricate | chewbbaca) | megahit |
fastqc_trimmomatic fastqc (spades pilon (
mlst | prokka | chewbbaca) | skesa pilon abricate))'
-o my_nextflow_pipelinewait, what?
FlowCraft
| Building features
Forks
Connect one component to multiple
Secondary channels
Connect non-adjacent components
Extra inputs
Inject user input data anywhere
Recipes
Curated and pre-assembled pipelines for specific needs
FlowCraft
| Live monitoring
Tracks Nextflow execution in real time:
FlowCraft
| Live reports
Dynamic generation of interactive report page
Multiple Raw Input Types
Not limited to paired-end FastQ or Fasta
Dynamic Input in Components
One component, multiple inputs
Expand Building Features
New merge operators
- Component that receives multiple input types
- Component that merges multiple inputs from different branches
FlowCraft
| The future
FlowCraft
| The team




Diogo N Silva
Tiago F Jesus
Inês Mendes
Bruno
Ribeiro-Gonçalves
Core developers

Advisors
Prof. Mário Ramirez

Prof. João A Carriço

Thank you for your attention
and happy pipeline building
Join the fun!



conda install flowcraftpip install flowcraftFCT PhD Grant SFRH/BD/129483/2017
Funding and Acknowledgements





