Predictability, computability, and stability for interpretable and reproducible data science
Karl Kumbier
Joint work with Bin Yu
Domain insights through supervised learning
Natural phenomena
Supervised learning
Domain insights
Prediction error
Interpretation error
The PCS framework: transparently evaluate human judgement calls
- Predictability: does my model capture external reality? (Popper, 1959; Allen, 1974; Stone, 1974; Breiman, 2001)
- Computability: can I tractably build/train my model? (Turing 1937; Hartmanis and Stearns, 1965)
- Stability: are my results consistent with respect to "reasonable" perturbations of the data/model? (Fisher, 1937; Popper 1959; Donoho et al., 2009; Yu, 2013; Stark 2018)
iterative Random Forest to identify genomic interactions
- Iteratively re-weighted Random Forests (Breiman, 20001; Amaratunga et al., 2008)
- Generalized random intersection trees (Shah and Meinshausen, 2014)
- Stability bagging (Breiman, 1996)
Joint work with Sumanta Basu, James B. Brown, and Bin Yu
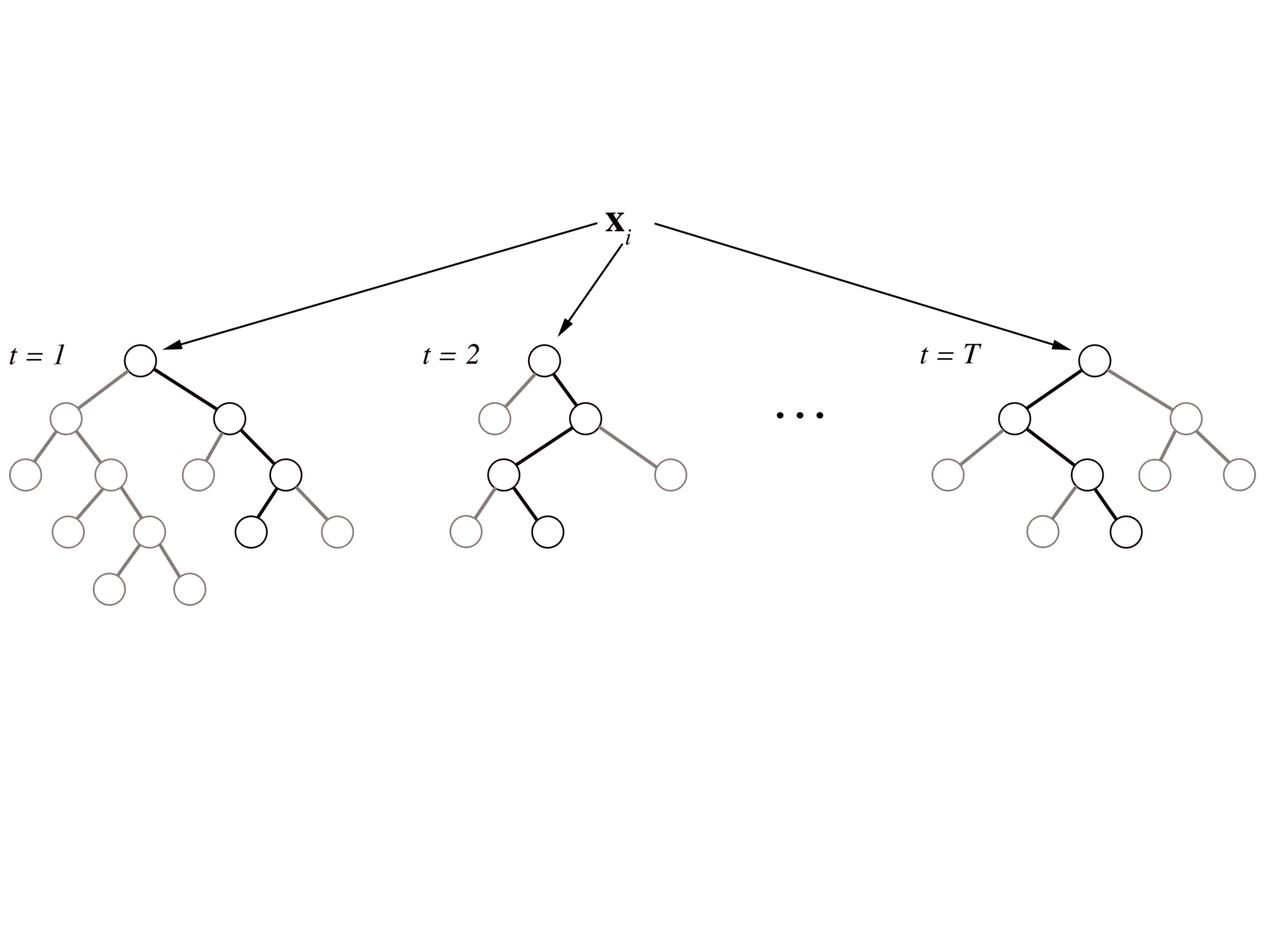
iterative Random Forests (iRF) build on PCS to identify genomic interactions in developing Drosophila embryos
iterative re-weighting stabilizes RF decision paths
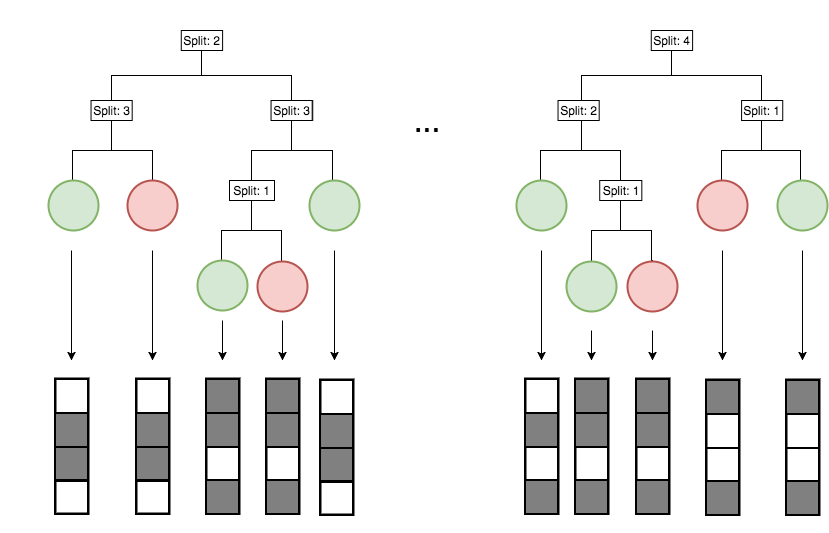
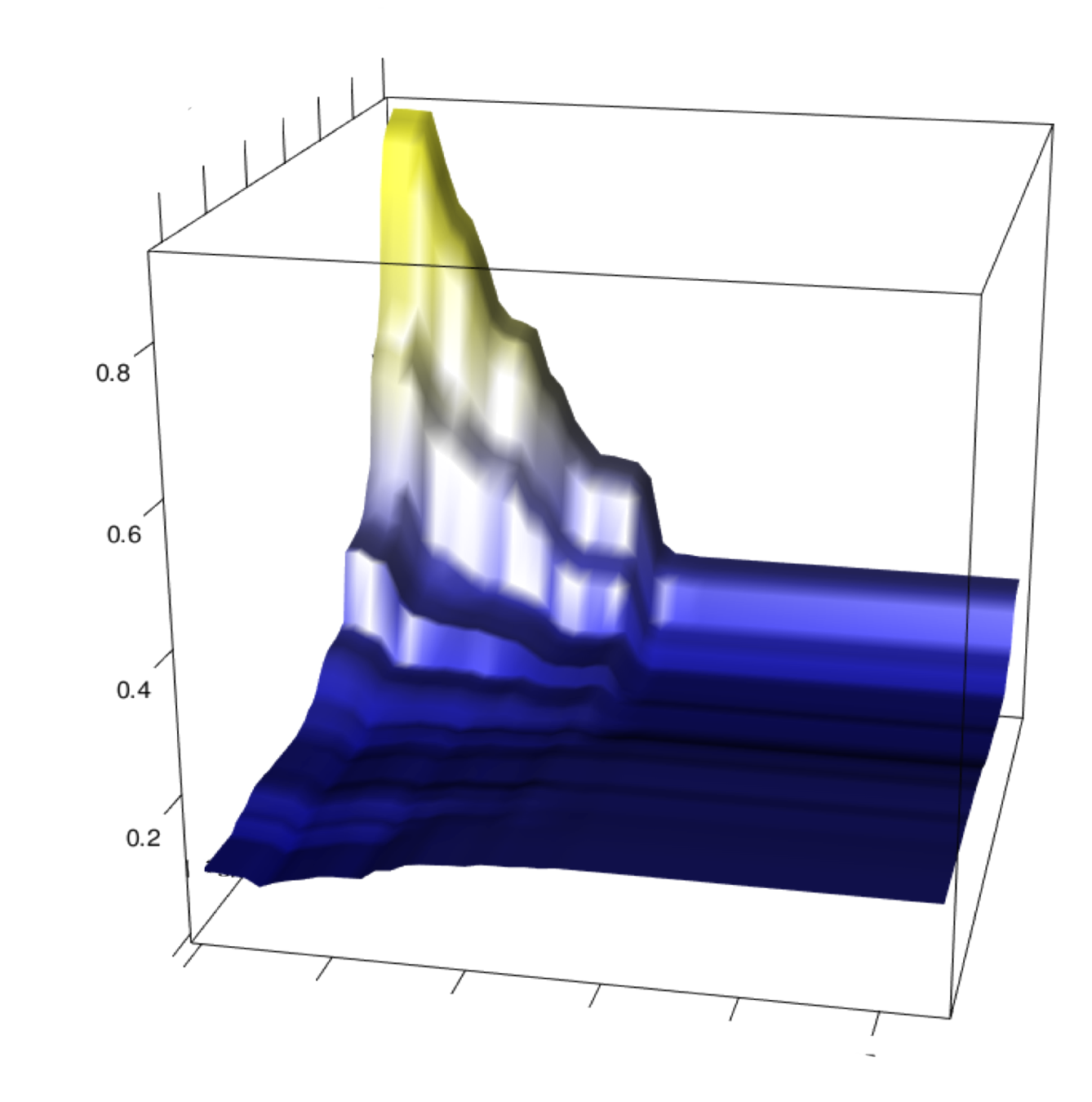
Sampling features based on their importance acts as soft regularization and maintains predictive accuracy of RF
Gini importance
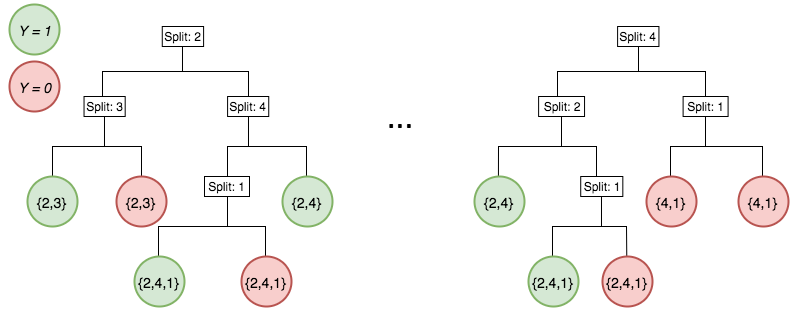
Generalized random intersection trees search for high-order interactions
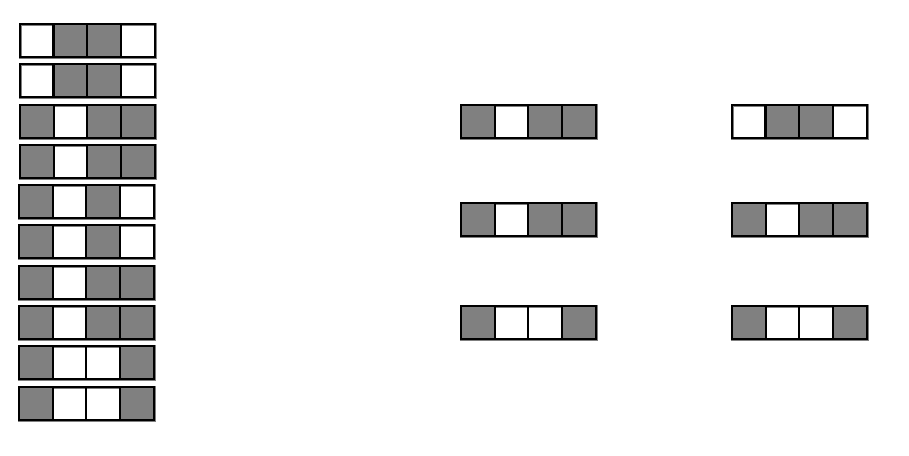
Features selected on RF decision paths
Intersect features on randomly selected decision paths to identify frequently co-occurring combinations
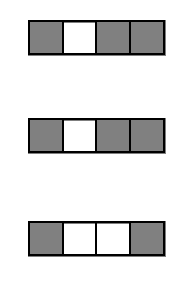
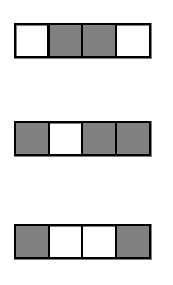
Bagging evaluates stability of interactions
Outer-layer bootstrap samples evaluate how consistently interactions are recovered across bootstrap perturbations of the data






Predicting early stage cancers
- 1817 patients (1005 cancer, 812 control)
- 8 cancer types:
ovary, liver, stomach, pancreas, lung, breast,
esophagus, and colorectum
-
61 genetic mutations and 39 protein biomarkers
Cancer detection: incorporating rules for better prediction
- iRF can extract predictive rules from a Random Forest
- Individual rules are interpretable (e.g. sparse, simulatable, modular) and can be screened based on domain constraints
Probability of cancer
Decision rule
iRF interaction


Rule-based predictions consistently improve predictive accuracy
Rule-based models successfully diagnose an additional 123/1000 patients under domain-specific constraints
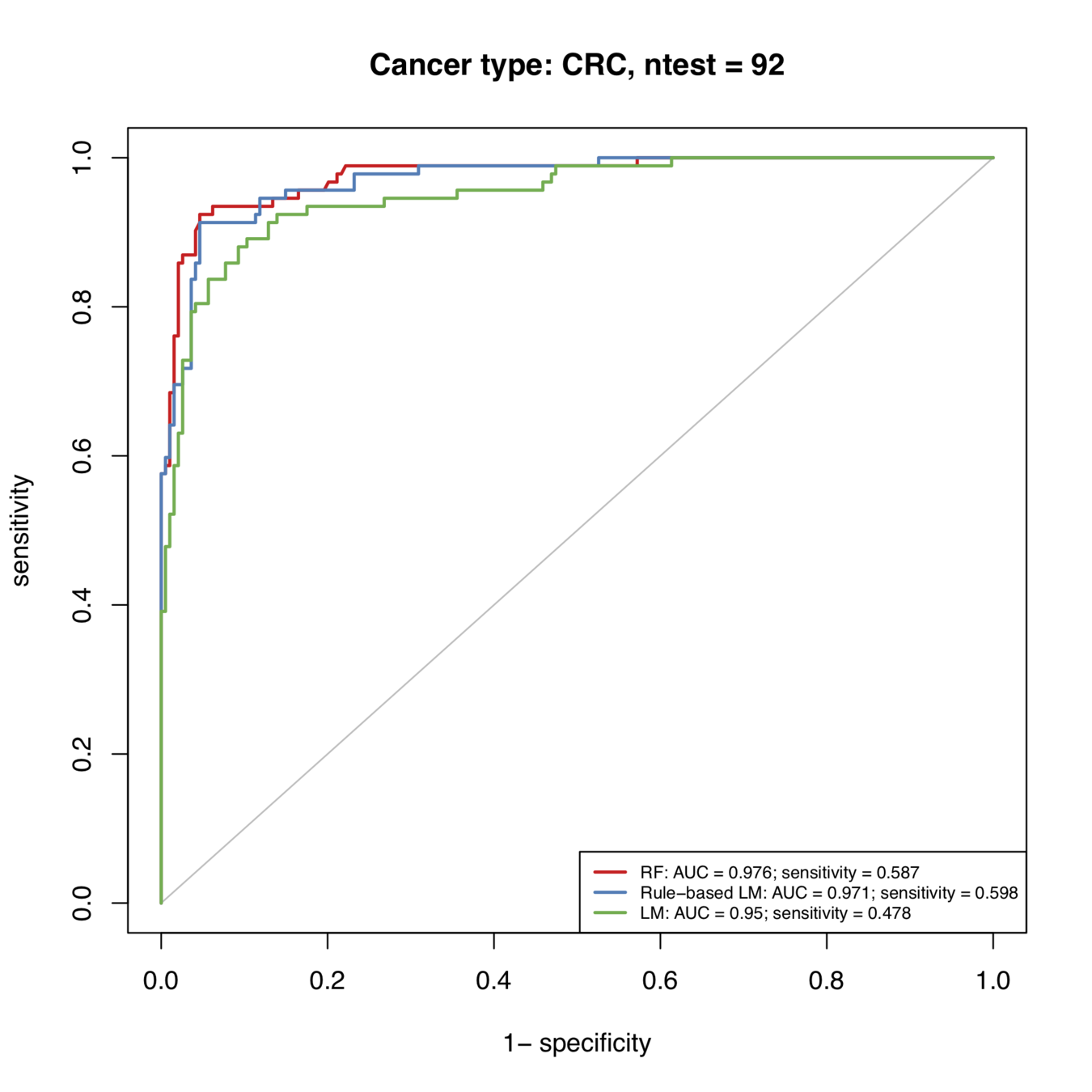
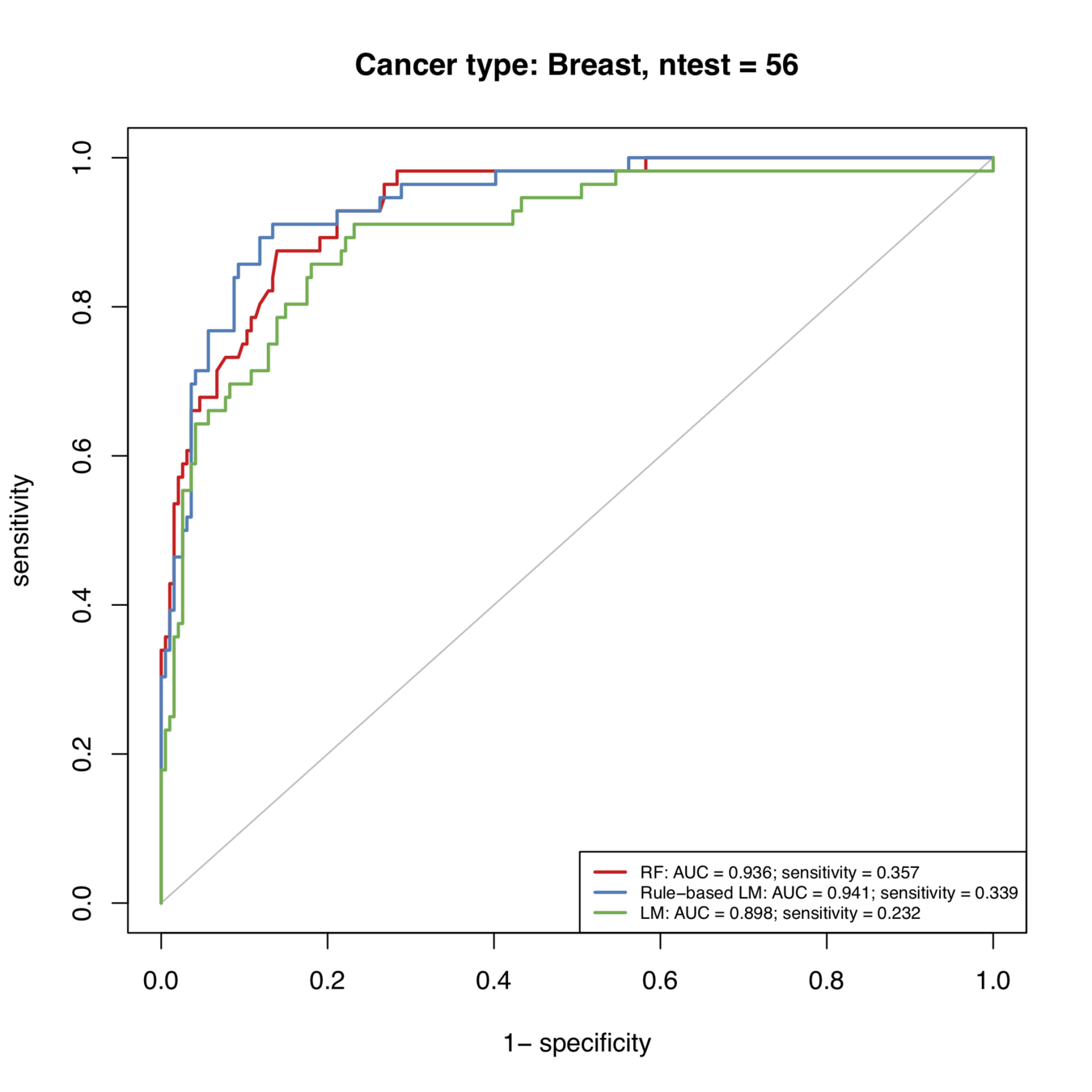
Rule-based predictions improve predictive accuracy over all cancer types
Additional 12 / 100 patients successfully diagnosed
Additional 11 / 100 patients successfully diagnosed
Summary
Ackowledgements
Defining interpretability
Interpretable machine learning: The extraction of relevant knowledge about domain relationships contained in data.
Relevant knowledge: provides insight for a particular audience into a chosen problem. These insights guide actions, discovery, and communication.
Encourage our model to learn interpretable relationships that can be [easily] read off by a practitioner.
Allow our model to learn complex relationships and develop methods to extract portions of these relationships.
Model based
Post hoc
Graphic for prediction accuracy vs interpretation accuracy
Model based interpretability
- Sparsity: how many components does my model have?
- Simulatability: can a human reasonably internalize the decision making process?
- Modularity: can the model be broken down into components that can be interpreted independently?
- Feature engineering
- Unsupervised learning
iRF builds in interpretability
iterative re-weighting encourages model to use sparse set of stable features
Recovered interactions represent predictive, simulatable "modules" (almost)
Post hoc interpretability
- Local interpretability: what relationship(s) does my model learn for an individual observation
- Global interpretability: what relationship(s) does my model learn across the entire dataset
Evaluating interpretability
- Interpretation accuracy: how well does my interpretation represent the model [image data relationship, prediction accuracy, interpretation accuracy]
- Interpretation stability: how consistent is my interpretation relative to reasonable perturbation(s) of the model/data
- Relevancy: does my interpretation address the task at hand without convoluting
Interpretability constraints in early-stage cancer prediction
Biomarkers known associations with cancer