Deep learning for
image data
Karl Kumbier
March 6, 2019
Deep learning: "end to end" representations
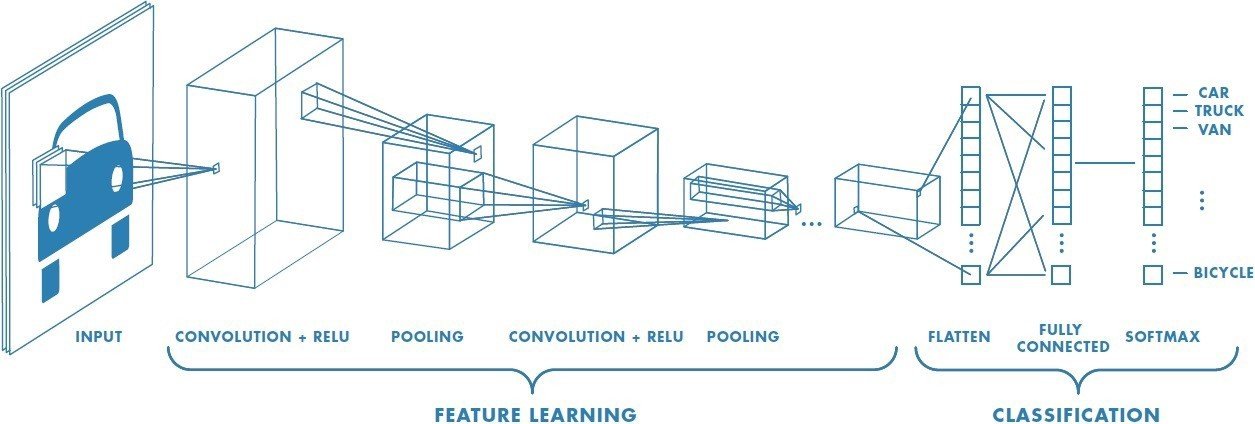
Convolutional neural network (CNN)
Fukushima (1980), LeCun et al. (1998), Krizhevsky (2012)
Deep learning: "end to end" representations

Convolutional neural network (CNN)
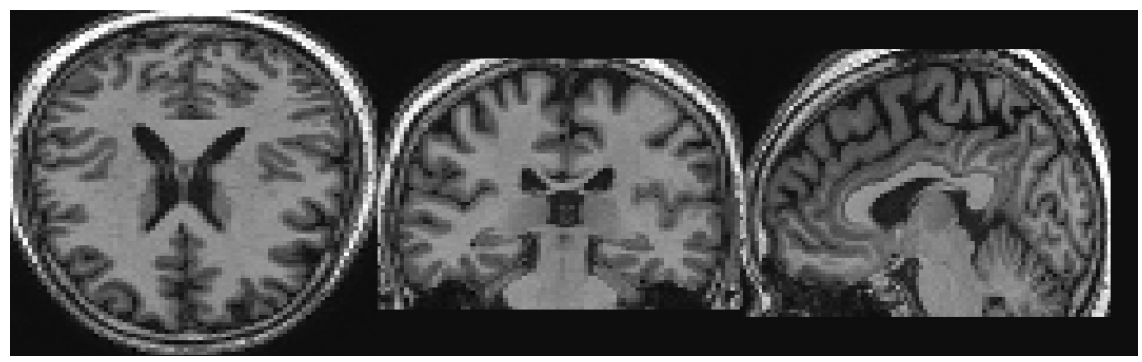
Classification
Payan and Montana (2015)
Fukushima (1980), LeCun et al. (1998), Krizhevsky (2012)
Deep learning: "end to end" representations

Convolutional neural network (CNN)

Classification
Payan and Montana (2015)
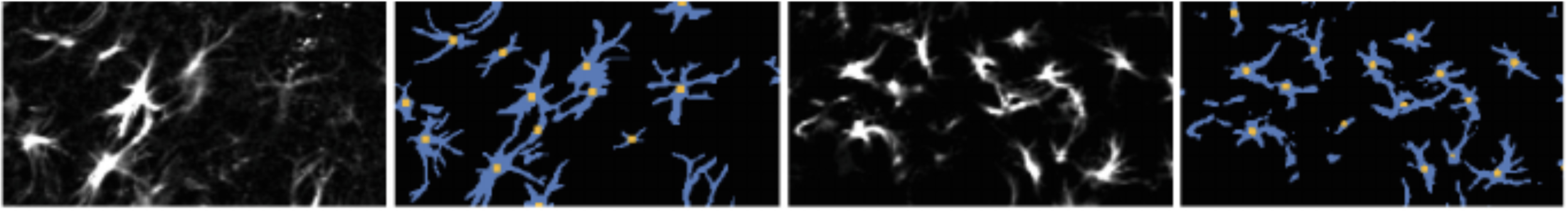

Segmentation
Yang et al. (2016)
Fukushima (1980), LeCun et al. (1998), Krizhevsky (2012)
Deep learning: "end to end" representations

Convolutional neural network (CNN)

Classification
Payan and Montana (2015)


Segmentation
Yang et al. (2016)
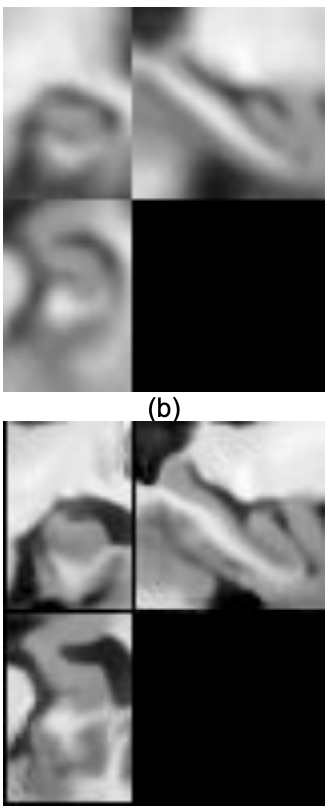

Registration
Li and Fan (2017)
Fukushima (1980), LeCun et al. (1998), Krizhevsky (2012)
Challenges for biomedical images
- Distinct imaging modalities (multi-channel, 3D, etc.)
- Limited availability of labeled data
Transfer learning
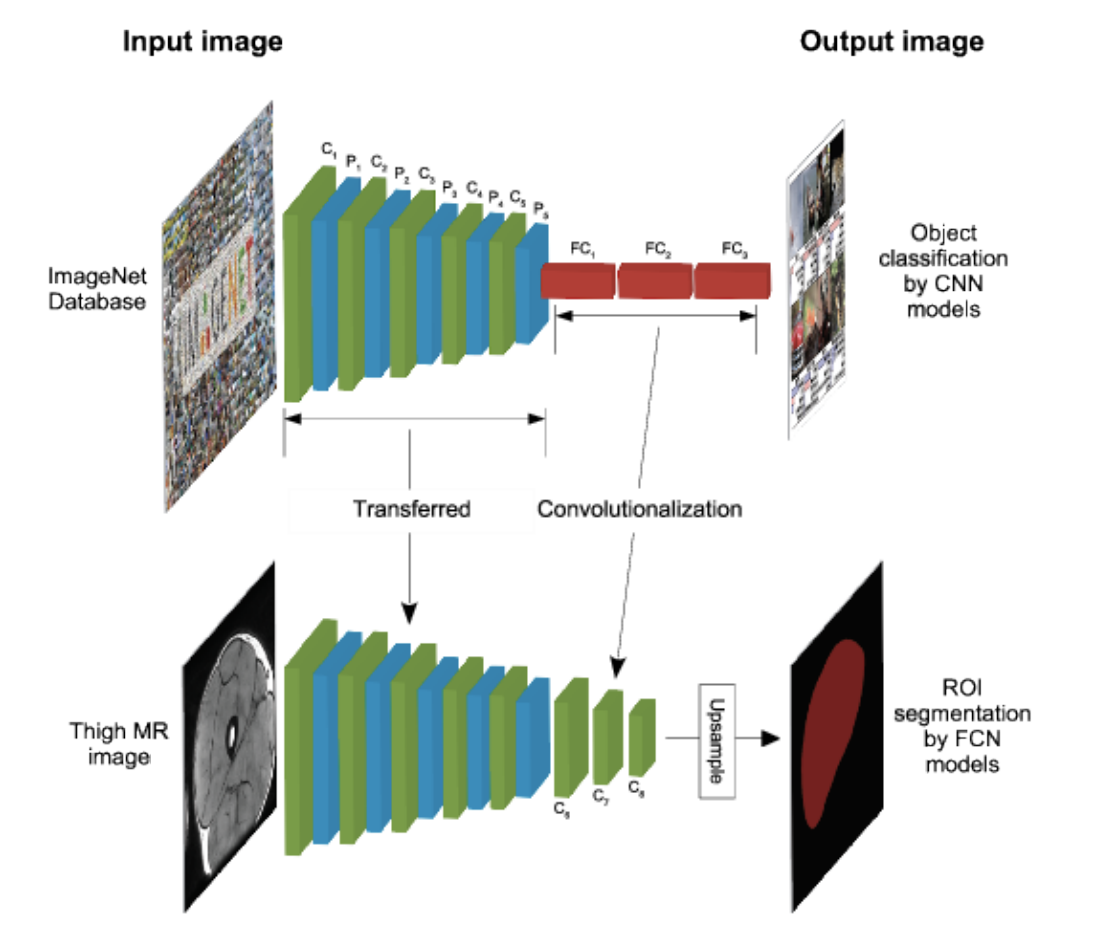
Antony et al. (2016), Kim et al. (2016)
- Distinct imaging modalities (multi-channel, 3D, etc.)
- Limited availability of labeled data
Challenges for biomedical images
Transfer learning

Antony et al. (2016), Kim et al. (2016)
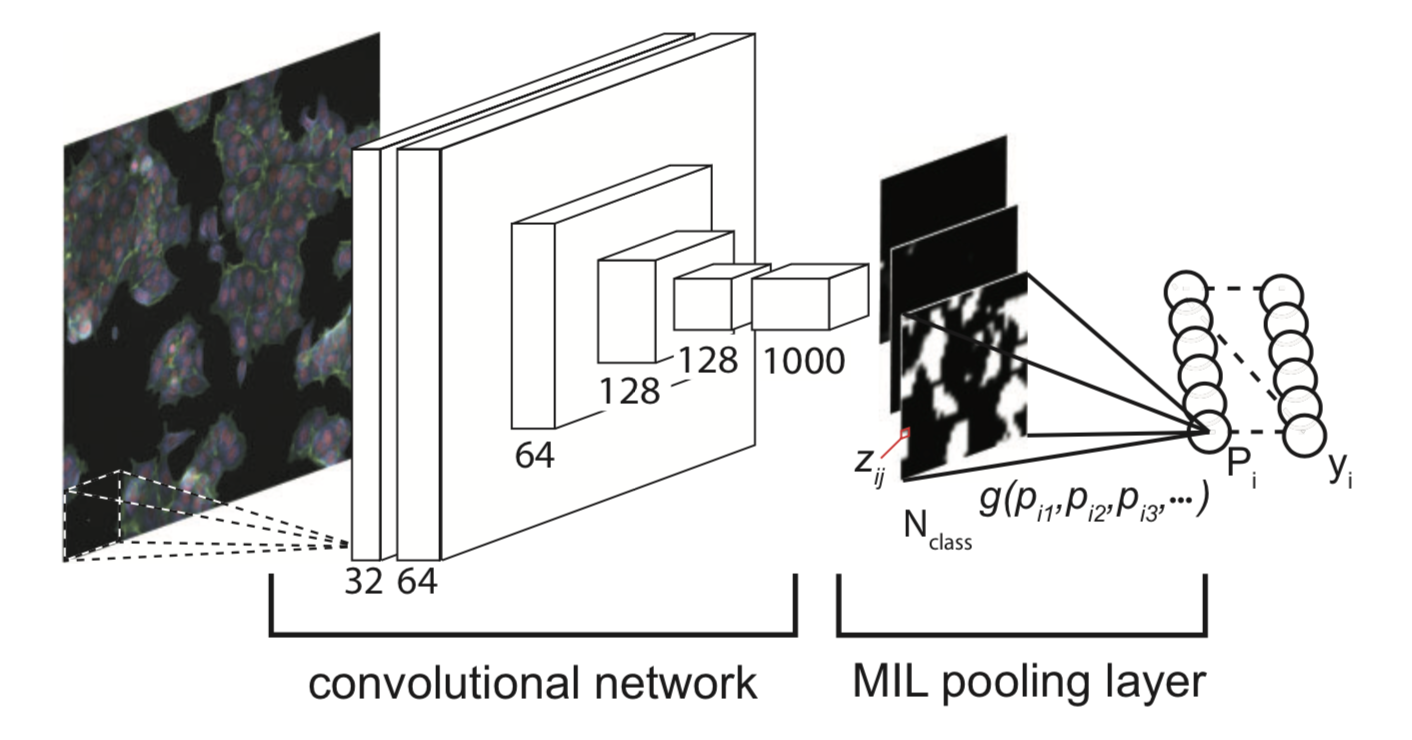
Problem formulation
Kraus et al. (2016)
- Distinct imaging modalities (multi-channel, 3D, etc.)
- Limited availability of labeled data
Challenges for biomedical images
Transfer learning
Data augmentation
Real

Antony et al. (2016), Kim et al. (2016)
Ronneberger et al. (2015), Uzunova et al. (2017)
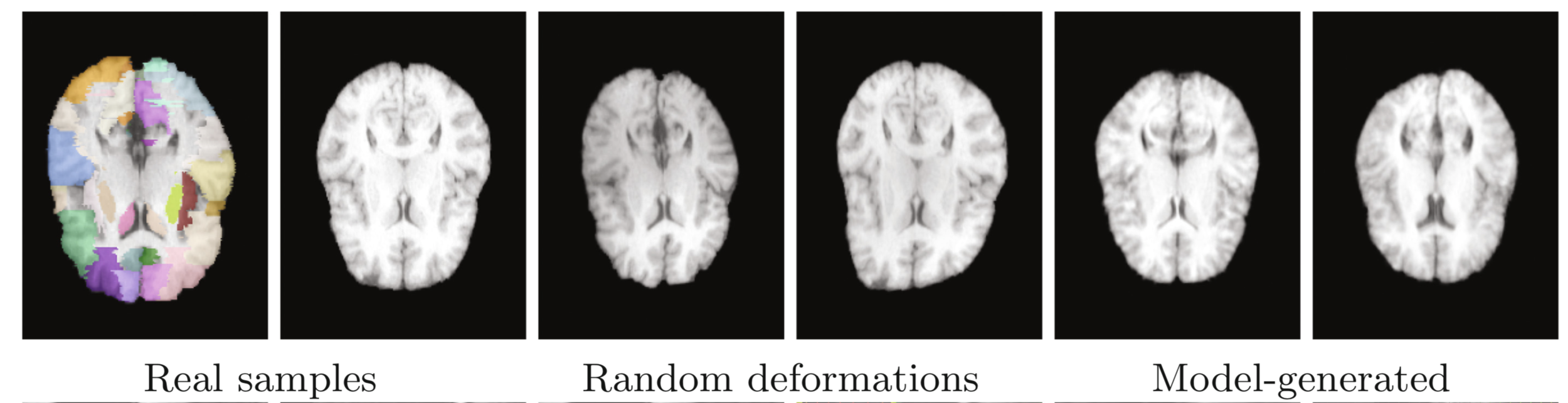


Deformed
Generated

Problem formulation
Kraus et al. (2016)
- Distinct imaging modalities (multi-channel, 3D, etc.)
- Limited availability of labeled data
Challenges for biomedical images
Deep learning in the lab
- Extracting high dimensional information from microscopy data
- Hypothesis generation/discovery from high dimensional data
- Improved automation