Lab Meeting
Leo Brueggeman
09/13/17
slides.com/leoo/lab-meeting
Systems Genetics of Neurodegeneration
Frauenchiemsee island, Chiemsee


Frauenchiemsee island, Chiemsee





Format
Isolation retreat
One week
25 young researchers (70% senior grad students, postdocs, residents, industry)
Represented countries: USA, Germany, Slovenia, France, UK, Australia, Sweden, Korea
Project focused
First 1/2 of week: 3-4 hrs of lectures per day + 4-5 hrs research time
Last 1/2 of week: Unscheduled group work w/ facilitator overview
2 Presentations + 1 'peer review' session of manuscript draft at end

Resources
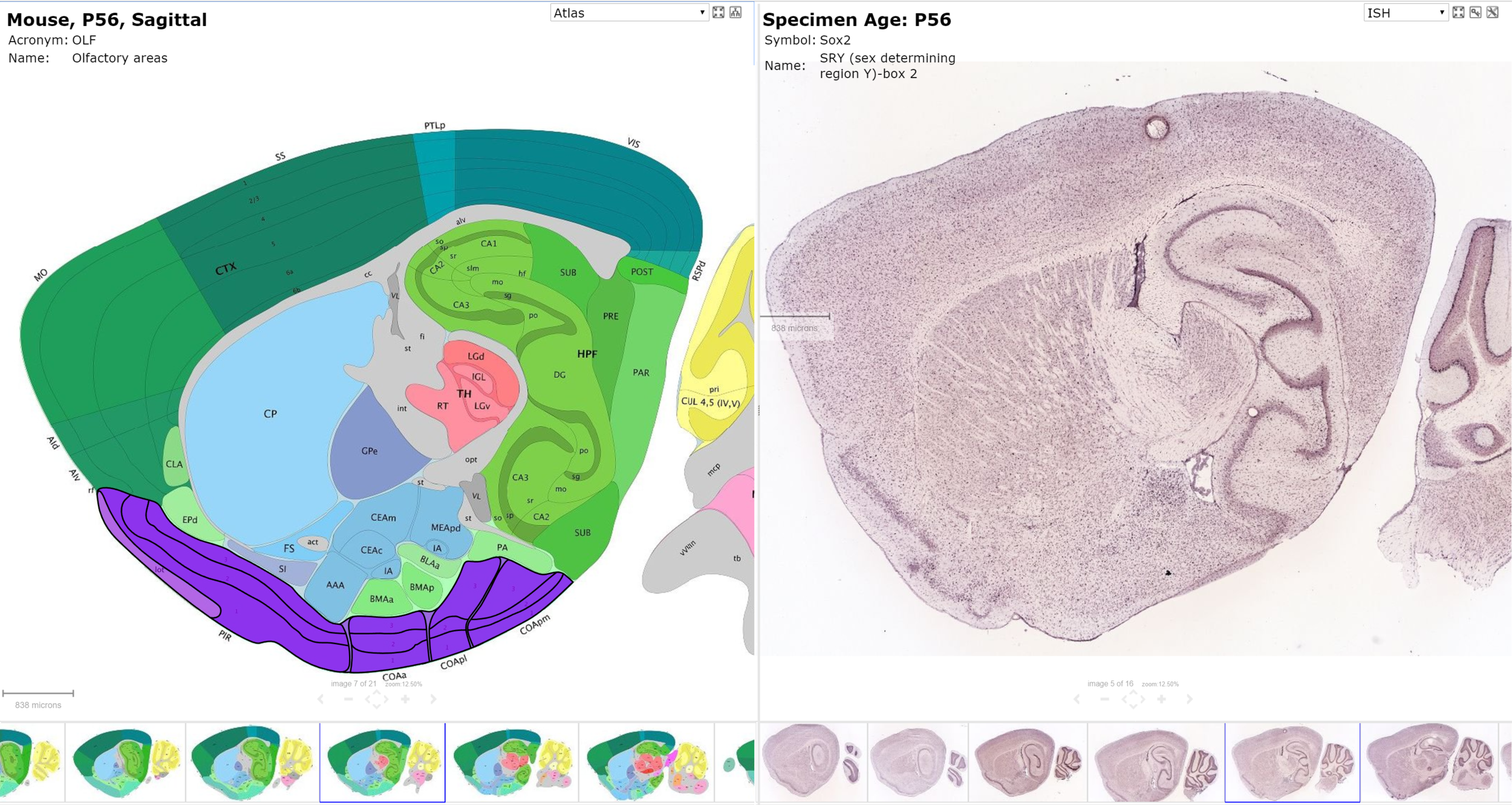
Allen Brain Institute
- Mouse Brain Atlas (in situ hybridization, 20k genes)
- Spinal cord atlas (ISHybrid, 17k genes, mouse, young & adult)
- Developing mouse brain atlas (microarray)
- Mouse connectivity atlas (see projections of cells w/in the brain, specific cell lines)
- BrainSpan
- Allen human brain atlas
- Gliobastoma
- Aging, Dementia, and TBI (100 individuals, 80-100 yo)
- Allen cell types database (electrophysiology, morphology, and rnaseq, mouse only for now)
- Allen brain observatory (response of cortex to mice doing visual behaviors)
Allen Human Brain Atlas
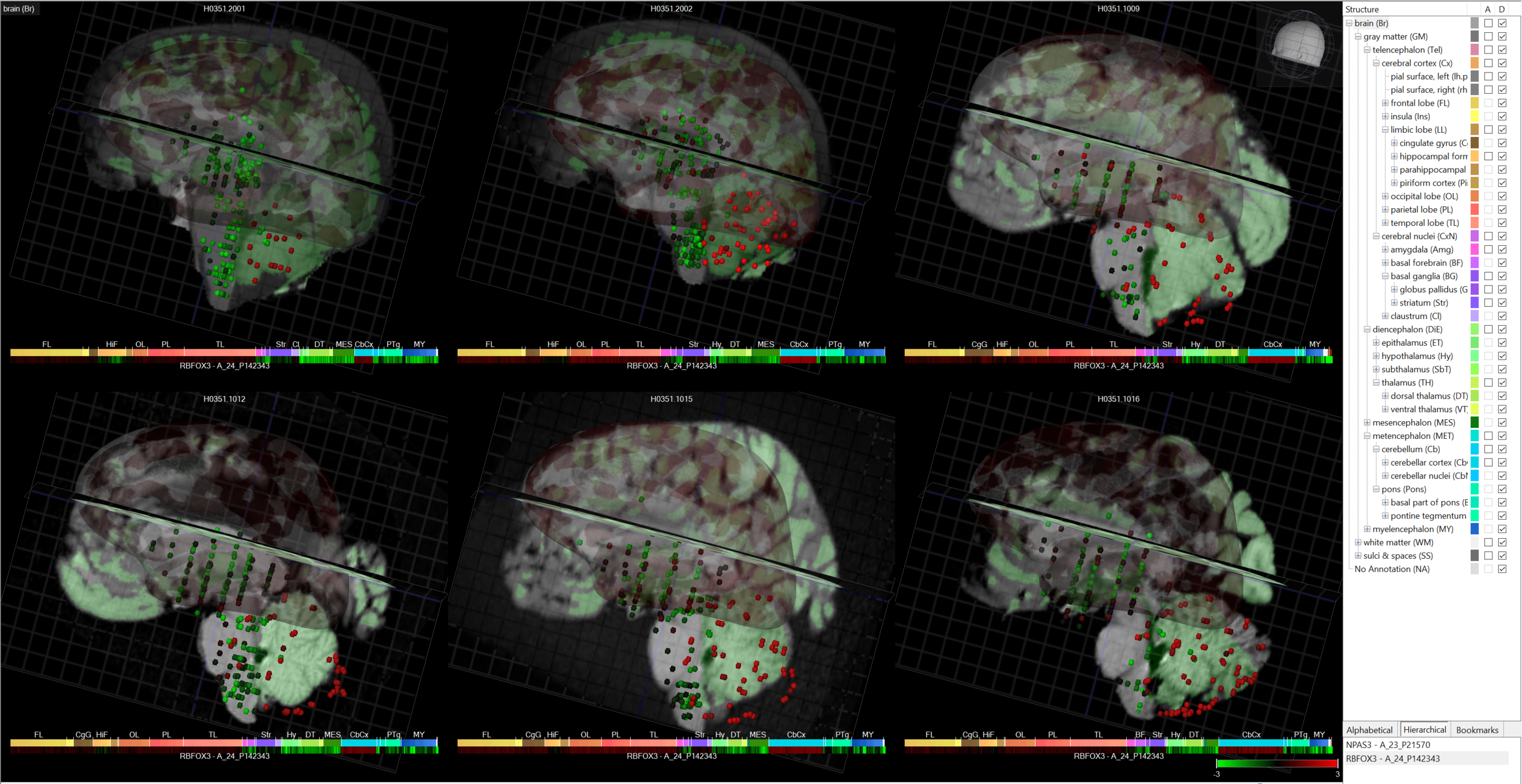
Allen Mouse Brain Atlas
BxD (genenetwork.org)

Genotypes
Phenotypes
genenetwork.org
Group Project
Nathan Skene. Postdoc in Sten Linnarsson's lab at Karolinska Institute (Sweden). Wet & dry lab research focused on single cell, brain



Annika Sommer, graduating PhD student from University of Erlangen-Nuremberg. Wet lab research on Parkinson's disease
Zoe Hanss, 3rd year PhD student at Luxembourg Center for Systems Biomedicine. Wet lab research on Parkinson's disease
Sara Redensek, 3rd year PhD student at University of Ljubljana, Slovenia. Wet lab research on Parkinson's disease
The team
Wet lab scientists work on PD
Last talk was about DJ1, a mitochondrial 'PARK' gene
Agree project going to be on PD
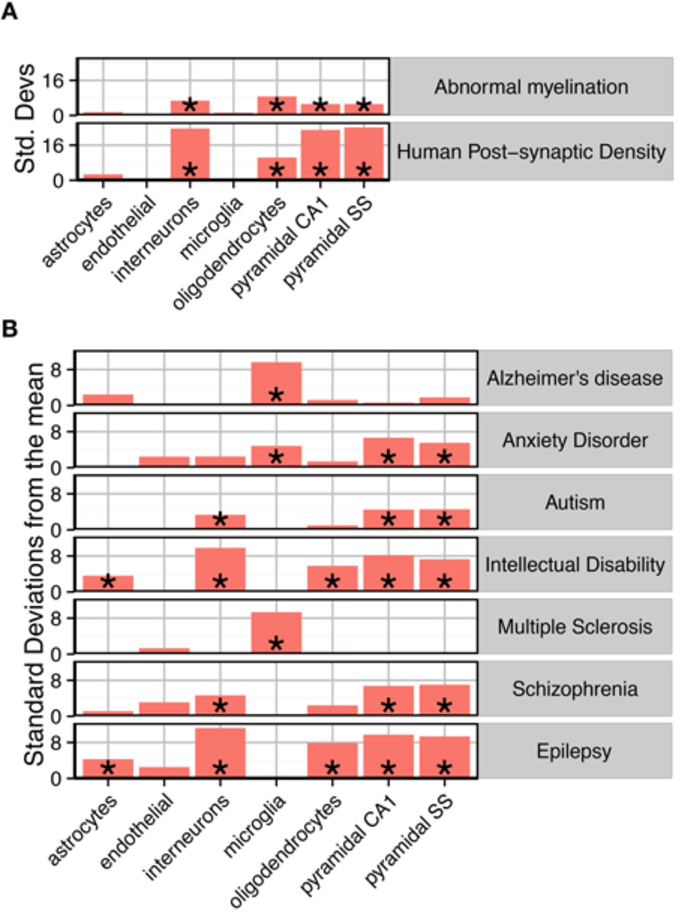
Does PD have an enrichment of mitochondrial genes?
| Source of dz genes | Association type |
|---|---|
| GWAS + familial genes | Causative |
| RNAseq from PD cases | Correlated |
Mitomap: 1000+ genes involved w/ mitochondrial fxn
hypergeometric test => no enr
Risk factors (aging, addiction protective), prodromal phase
Maybe 'aging' mt genes enriched?
Broader: what organelles aff in PD - use subcellular location
Broader: what cell types aff in PD - use single cell data. Prodromal
Broader: what brain regions aff in PD - use Allen Inst data
Project Conception Map
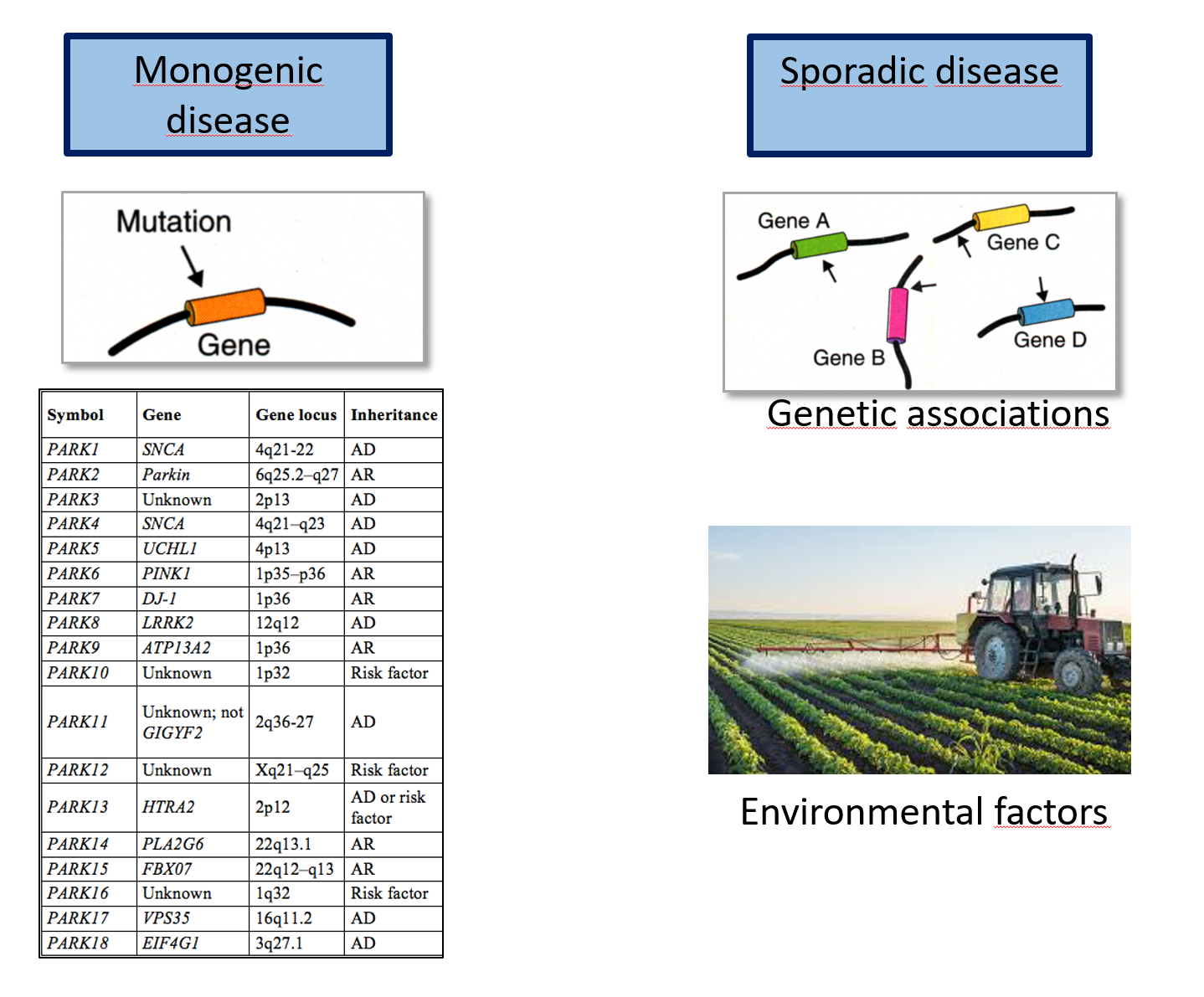
Etiology of PD
| Cell type x1 | ... | Cell type xn | |
|---|---|---|---|
| Gene y1 expr | 5 | 0 | 0 |
| ... | 2 | 2 | 2 |
| Gene yn expr | 0 | 1 | 4 |
Approach
| Core idea: |
|---|
| Cell type x1 | ... | Cell type xn | |
|---|---|---|---|
| Gene y1 spec | 5/5+0 = 1 | 0/0+5 = 0 | 0/0+5 = 0 |
| ... | 2/6 = .33 | 2/6 = .33 | 2/6 = .33 |
| Gene yn spec | 0/4 = 0 | 1/4 = .25 | 3/4 = .75 |
Approach
| Core idea: |
|---|
| Cell type x1 | ... | Cell type xn | |
|---|---|---|---|
| Gene y1 spec | 5/5+0 = 1 | 0/0+5 = 0 | 0/0+5 = 0 |
| ... | 2/6 = .33 | 2/6 = .33 | 2/6 = .33 |
| Gene yn spec | 0/4 = 0 | 1/4 = .25 | 3/4 = .75 |
| GWAS result: gene a, gene b, ... |
|---|
| Cell type x1 | ... | Cell type xn | |
|---|---|---|---|
| Gene a | .8 | .1 | .1 |
| Gene b | .7 | .2 | .1 |
| ... | .9 | .05 | .05 |
| Sum of spec | 2.4 | .35 | .25 |
|---|
| P value: |
|---|
Permute random sets of genes from background genes, building a distribution of specificity scores
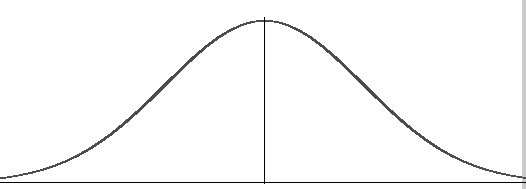
Example of Approach