Mikel Egaña Aranguren
Candidato Ayudante Doctor
Dpto. Ciencia de la Computación e Inteligencia Artificial UPV/EHU
http://mikel-egana-aranguren.github.io
mikel.egana.aranguren@gmail.com
Meritos preferentes
Estancias en centros diferentes a UPV/EHU














Experiencia laboral y becas
Experiencia laboral
(2015 - Presente) Analista Eurohelp Consulting, S.L.
(2014-2015) Investigador UPV/EHU
(2011-2014) Investigador UPM
(2009) Investigador UM
(2006) Investigador pre-doc VIB, Bélgica
(2005-2009) Doctorado Universidad de Manchester, UK
Becas

Marie Curie Cofund Postdoc (UE): 2011 - 2014
Marie Curie EST Predoc (UE): 2006
EPSRC/Universidad de Manchester Doctorado (UK): 2005 - 2008
Erasmus (UE, BBK), Canterbury Christ Church, UK: 2001
Torres Quevedo (MINECO): 2015 - 2018





Méritos de investigación
Visitas de investigación
2005 European Bioinformatics Institute (UK)
Financiado por Network of Excellence on Semantic Interoperability and Data Mining in Biomedicine (EU)
Desarrollo Gene Ontology
Participación en proyectos
2016 – 2020 (Ongoing). REPLICATE: Renaissance of Places with Innovative Citizenship And TEchnology (Project 691735). EU. (Consortium: 24M EUR; Eurohelp: 300.000 EUR)
2015 – 2018 (Ongoing). Linking Open Domains, Plataforma para la generación de datos enlazados (LODGen). Ministerio de Industria, Energia y Turismo, Spain (TSI-100105-2015-0012). (40.000 EUR)
2016 – 2017 (Ongoing). Servicios OpenLinkedData (Linked Data implementation of Open Data Euskadi). Basque Government, Spain. (90.000 EUR)
2015 Analista en proyecto ENGIMU (Enlazando Gipuzkoa con el Mundo), financiado por Gipuzkoako Foru Aldundia, 40.000€

Publicaciones en revistas JCR
Aranguren, M., Bechhofer, S., Lord, P., Sattler, U., and Stevens, R. (2007). Understanding and using the meaning of statements in a bio-ontology: recasting the Gene Ontology in OWL. BMC bioinformatics, 8(1):57
[Factor Impacto: 2.58]

Stevens, R., Egaña Aranguren, M., Wolstencroft, K., Sattler, U., Drummond, N., Horridge, M., and Rector, A. (2007). Using OWL to model biological knowledge. International Journal of Human-Computer Studies, 65(7):583–594
[Factor Impacto: 1.293]
Egaña Aranguren, M., Wroe, C., Goble, C., and Stevens, R. (2008). In situ migration of handcrafted ontologies to reasonable forms. Data & Knowledge Engineering, 66(1):147–162
[Factor Impacto: 1.115]
Antezana, E., Egaña, M., De Baets, B., Kuiper, M., and Mironov, V. (2008b). ONTO-PERL: an API for supporting the development and analysis of bio-ontologies. Bioinformatics, 24(6):885
[Factor Impacto: 4.981]
Aranguren, M., Antezana, E., Kuiper, M., and Stevens, R. (2008a). Ontology Design Patterns for bio-ontologies: a case study on the Cell Cycle Ontology. BMC bioinformatics, 9(Suppl 5):S1
[Factor Impacto: 2.58]

Antezana, E., Blondé, W., Egaña, M., Rutherford, A., Stevens, R., De Baets, B., Mironov, V., and Kuiper, M. (2009a). BioGateway: a semantic systems biology tool for the life sciences. BMC bioinformatics, 10(Suppl 10):S11
[Factor Impacto: 2.58]
Antezana, E., Egaña, M., Blondé, W., Illarramendi, A., Bilbao, I., De Baets, B., Stevens, R., Mironov, V., and Kuiper, M. (2009b). The Cell Cycle Ontology: an application ontology for the representation and integrated analysis of the cell cycle process. Genome Biology, 10(5):R58
[Factor Impacto: 10.8]

Minarro-Gimenez, J., Egana-Aranguren, M., Villazon-Terrazas, B., and FernandezBreis, J. (2012). Publishing Orthology and Diseases Information in the Linked Open Data Cloud. Current Bioinformatics, 7(3):255–266
[Factor Impacto: 0.971]
Mironov, V., Antezana, E., Egaña, M., Blondé, W., De Baets, B., Kuiper, M., and Stevens, R. (2011). Flexibility and utility of the Cell Cycle Ontology. Applied Ontology, 6(3):247–261
[Factor Impacto: 0.615]
Miñarro-Gimenez, J., Aranguren, M., Béjar, R., Fernández-Breis, J., and Madrid, M. (2011). Semantic integration of information about orthologs and diseases: The OGO system. Journal of biomedical informatics, 44:1020–1031
[Factor Impacto: 2.126]
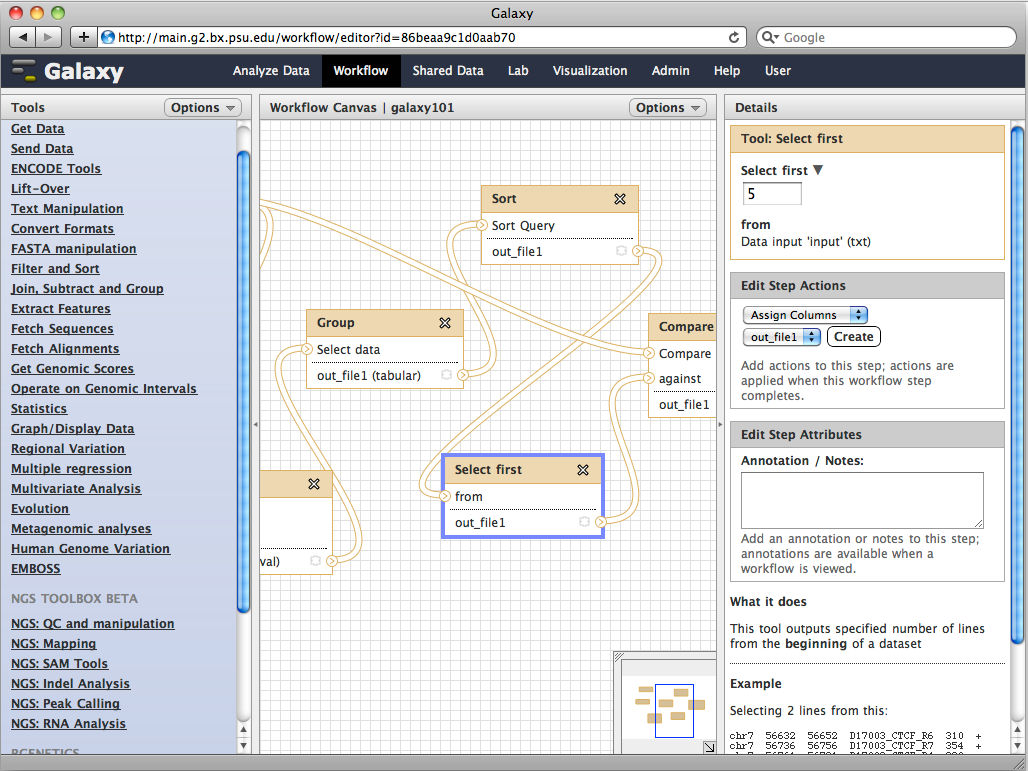
Egaña Aranguren, M., Fernández-Breis, J. T., Antezana, E., Mungall, C., Rodríguez González, A., and Wilkinson, M. D. (2013). OPPL-Galaxy, a Galaxy tool for enhancing ontology exploitation as part of bioinformatics workflows. Journal of biomedical semantics, 4(1):2
[Factor Impacto: 2.24]
Duque-Ramos, A., Fernández-Breis, J. T., Iniesta, M., Dumontier, M., Egaña Aranguren, M., Schulz, S., Aussenac-Gilles, N., and Stevens, R. (2013). Evaluation of the OQuaRE framework for ontology quality. Expert Systems with Applications, 40(7):2696–2703.
[Factor Impacto: 2.26]
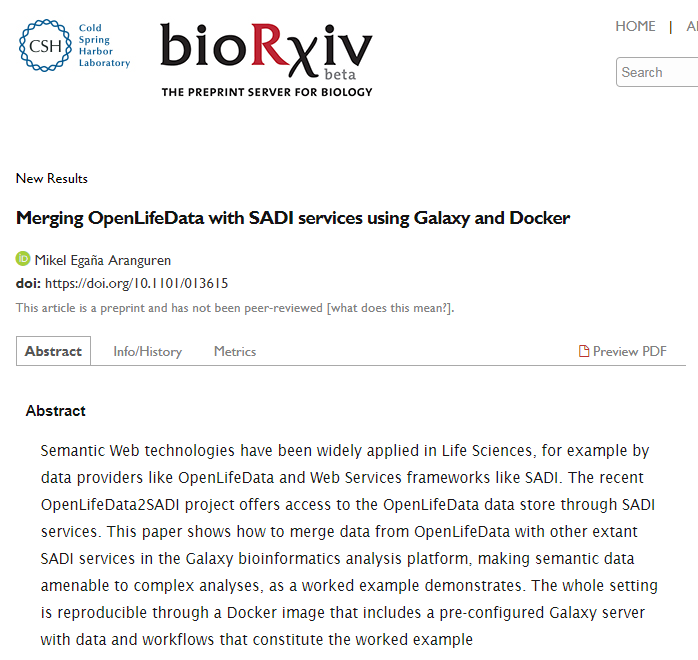
Aranguren, M. E., González, A. R., and Wilkinson, M. D. (2014). Executing SADI services in Galaxy. Journal of Biomedical Semantics, 5(1):42+.
[Factor Impacto: 2.26]
José Antonio Miñarro Giménez, Mikel Egaña Aranguren, Boris Villazón Terrazas, and Jesualdo Tomás Fernández Breis (2014). Translational research combining orthologous genes and human diseases with the OGOLOD dataset. Semantic Web Journal, 5(2):145–149
[Factor Impacto: 1.786]
González, A. R., Callahan, A., Toledo, J. C., García, A., Aranguren, M. E., Dumontier, M., and Wilkinson, M. D. (2014a). Automatically exposing OpenLifeData via SADI semantic Web Services. Journal of Biomedical Semantics, 5(1):46+
[Factor Impacto: 2.26]
Pawluczyk, M., Weiss, J., Links, M. G., Aranguren, M. E., Wilkinson, M. D., and Egea-Cortines, M. (2015). Quantitative evaluation of bias in PCR amplification and Next Generation Sequencing derived from metabarcoding samples. Analytical and Bioanalytical Chemistry, 407(7):1841–1848
[Factor Impacto: 3.436]
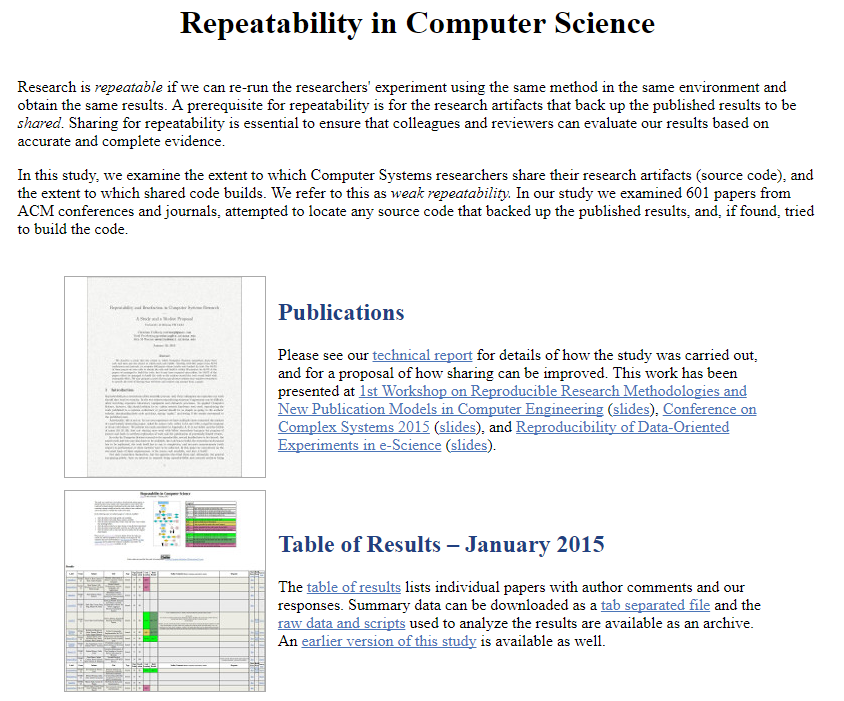
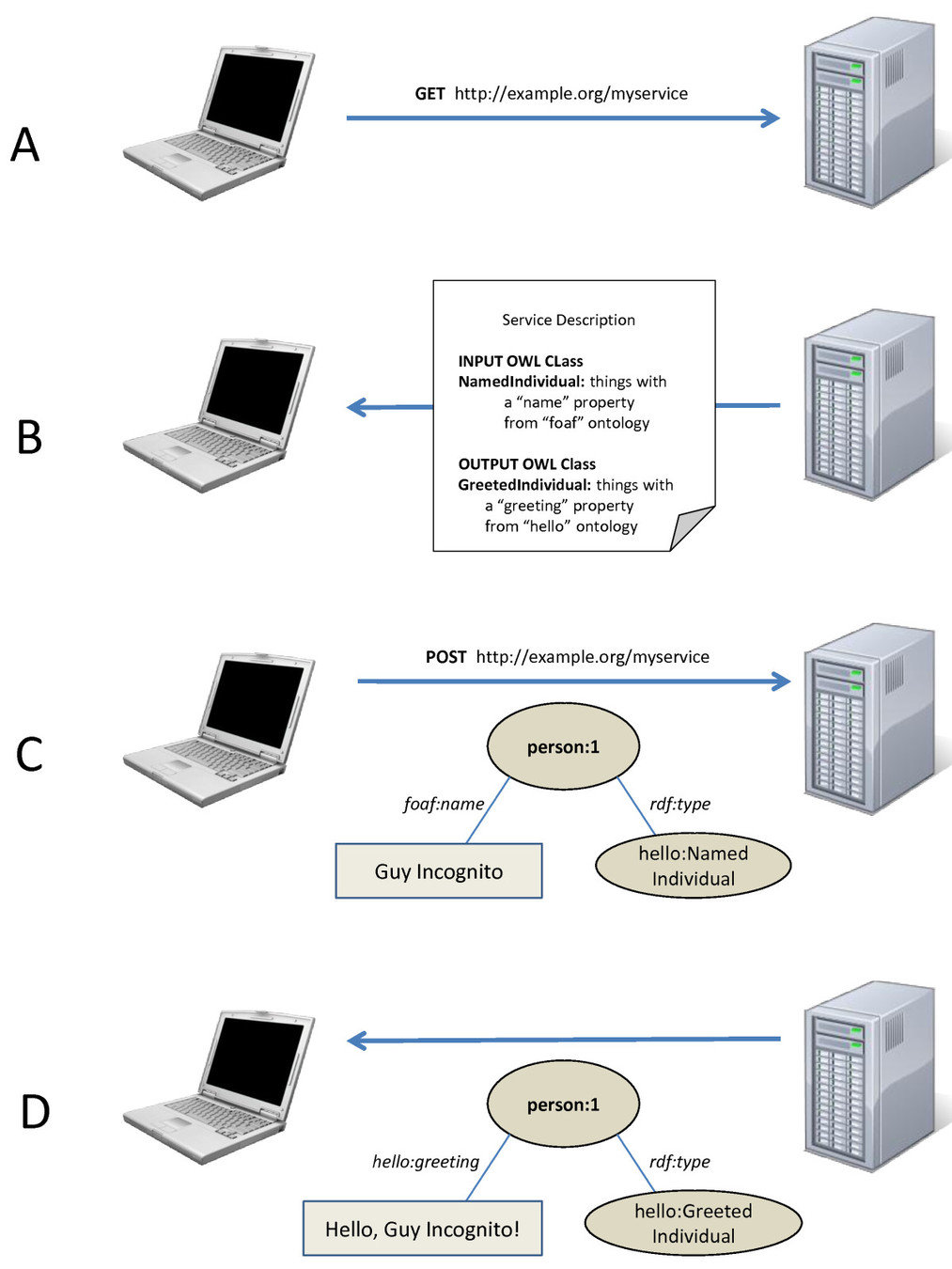
Mikel Egaña Aranguren, Mark D. Wilkinson (2015). Enhanced reproducibility of SADI Web service workflows with Galaxy and Docker. GigaScience, 4(59)
[Factor Impacto: 7.46]







3.- Semantics: what the data means
2.- Functional: what I did with the data
1.- Computational: how I did it
Analytical and Bioanalytical Chemistry (1), GigaScience (1), Genome Biology (1)
BMC Bioinformatics (3), Bioinformatics (1), Current Bioinformatics (1), Journal of Biomedical Informatics (1)
Journal of Biomedical Semantics (3)
Semantic Web Journal (1), Internationl Journal of Human Computer Studies (1), Data and Knowledge Engineering (1), Applied Ontology (1), Expert Systems With Applications (1)
Capítulos en libros
Aranguren, M., Stevens, R., Antezana, E., Fernández-Breis, J.T., Kuiper, M., and Mironov, V. (2010). Technologies and Best Practices for Building Bio-Ontologies. In Knowledge-Based Bioinformatics, volume Gil Alterovitz and Marco Ramoni (Eds.), pages 67–86. Wiley Online Library
Editor
Mikel Egaña Aranguren, Jesualdo Tomás Fernández Breis, and Michel Dumontier (2014). Special issue on Linked Data for Health Care and the Life Sciences. Semantic Web Journal, 5(2):99–100
Ponencias en congresos
González, A. R., Romero, M. M., Aranguren, M. E., and Wilkinson, M. D. (2014b). Nanopublishing clinical diagnoses: tracking diagnostic knowledge base content and utilization. In 27th International Symposium on Computer-Based Medical Systems (CBMS), pages 335–340
Iglesias, A. R., Aranguren, M. E., González, A. R., and Wilkinson, M. D. (2013). Plant Pathogen Interactions Ontology (PPIO). In Rojas, I. and Guzman, F. M. O., editors, IWBBIO, pages 695–702. Copicentro Editorial
Aranguren, M., Fernández-Breis, J., and Antezana, E. (2011). OPPL-Galaxy: enhancing ontology exploitation in galaxy with OPPL. In Proceedings of the 4th International Workshop on Semantic Web Applications and Tools for the Life Sciences, pages 12–19. ACM
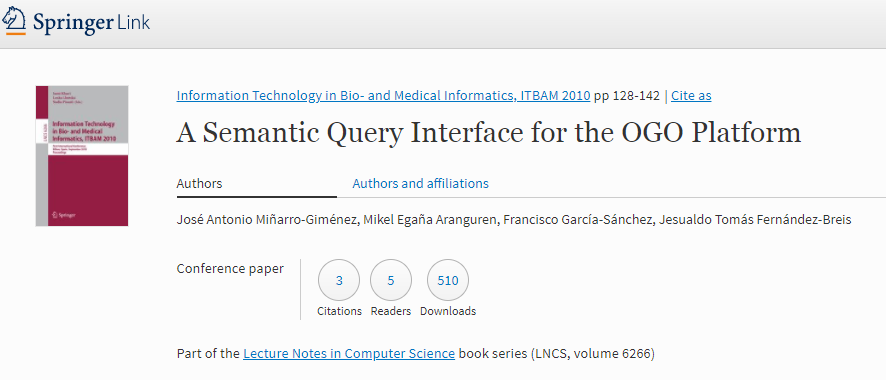
Miñarro-Giménez, J., Aranguren, M., García-Sánchez, F., and Fernández-Breis, J. (2010). A semantic query interface for the OGO platform. In Information Technology in Bio-and Medical Informatics, ITBAM 2010, pages 128–142. Springer
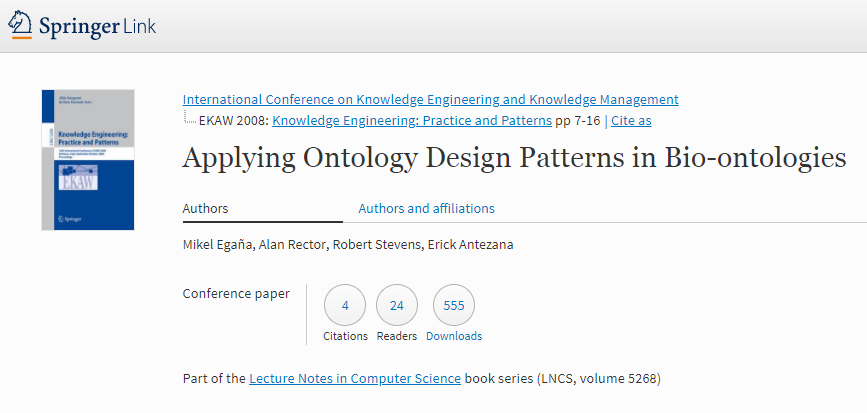
Egaña, M., Rector, A., Stevens, R., and Antezana, E. (2008). Applying ontology design patterns in bio-ontologies. In Gangemi, A. and Euzenat, J., editors, Knowledge Engineering: Practice and Patterns, volume 5268 of Lecture Notes in Computer Science, pages 7–16. Springer
Charlas invitadas
Los Datos Enlazados y la Web Semántica. Tikitalka, VE Interactive, Spain, 2016
Building reasonable biomedical ontologies for a Life Sciences Semantic Web. 3S (Systems, Synthetic, and Semantic) Biology summer school. CIBIO (Centre for Integrative Biology), University of Trento, Italy, 2014
Linked Data for Functional Genomics. NTNU, Trondheim, Norway, 2011
Aplicación de la Web Semántica en Biología Molecular. Universidad de Deusto, Facultad de Ingenieria, Spain, 2010
Aplicación de la Web Semántica en Bioinformática. UM, Facultad de Informática, Spain, 2008
Métodos y resultados actuales en Bioinformática: know-how y know-what de las redes tecnocientíficas en Bioinformática. EHU, Facultad de Filosofia, Spain, 2004
Revisor
2015 BMC Medical Informatics and Decision Making
2013 PeerJ
2013 Data and Knowledge Engineering (DKE)
2012 BMC Bioinformatics
2012 Journal of Biomedical Informatics (JBI)
2012 Computational and Mathematical Methods in Medicine (CMMM)
2012 Journal of Medical Systems (JOMS)
2012 Journal of Biomedical Semantics (JBS)
2011 Semantic Web Journal (SWJ)
2011 Journal of Research and Practice in Information Technology (JRPIT)
Comité científico
2017 Semantic Web Solutions for Large-Scale Biomedical Data Analytics (SeWeBMeDA)
2015 CAEPIA
2012 Managing Interoperability and compleXity in Health Systems, in conjunction with the ACM International Conference on Information and Knowledge Management
2012 Joint Workshop on Semantic Technologies Applied to Biomedical Informatics and
Individualized Medicine (SATBI + SWIM 2012), in conjunction with International Semantic Web Conference (ISWC)
2011 Managing Interoperability and compleXity in Health Systems, in conjunction with the ACM International Conference on Information and Knowledge Management
2011 Knowledge Capture (K-CAP)
2011 Semantic Applied Technologies on Biomedical Informatics (SATBI 2011), in
conjunction with the ACM International Conference on Bioinformatics and Computational Biology
2008 ONTORACT
Métricas de producción científica
H-index (Google Scholar): 14
H-index (Scopus): 8
Citas totales (Google Scholar): 717
Perfiles de interés
GitHub: http://github.com/mikel-egana-aranguren
Google Scholar: http://scholar.google.com/citations?user=JsMMKnoAAAAJ
Scopus: http://www.scopus.com/authid/detail.url?authorId=16038705500
[Más información en https://mikel-egana-aranguren.github.io]
Meritos docentes y de gestión
Experiencia docente
Postgrado
2017 Life Sciences Semantic Web. MSc Bioinformatics, UM. Spanish
2016 Life Sciences Semantic Web. MSc Bioinformatics, UM. Spanish
2015 Life Sciences Semantic Web. MSc Bioinformatics, UM. Spanish
2014 Galaxy tutorial. Erasmus mundus MSc in Marine Environment and resources, UPVEHU. English
2014 Life Sciences Semantic Web. MSc Bioinformatics, UM. Spanish
2013 Life Sciences Linked Data. MSc Bioinformatics, UM. Spanish
2011 Web Ontology Language (OWL), Artificial Intelligence MSc (UPM). English
Grado
2012 OWL, as part of ATHENS course (UPM). English
2012 OWL, as part of ATHENS course (UPM). English
2011 OWL, as part of ATHENS course (UPM). English
2011 OWL/Description Logics, as part of the Artificial Intelligence course (UPM). Spanish
Misc.
2016 Linked Open Data tutorial. EJIE. Spanish
2015 Linked Open Data tutorial. IZFE (Informatika Zerbitzuen Foru Elkartea, Gipuzkoa). Spanish
2014 Semantic biology tutorial: Use of Semantic Web resources for knowledge discovery. 3S (Systems, Synthetic, and Semantic) Biology summer school. CIBIO (Centre for Integrative Biology), University of Trento, Italy. English
2013 Introductory talk on bioinformatics for high school students visiting the CBGP. Spanish
2013 Galaxy tutorials at CBGP. English and spanish
2011 Populous tutorial at SWAT4LS (London, UK), English
2005-2008 OWL tutorials for biologists (University of Manchester, UK). English
Tribunales doctorado
2014 Alejandro Rodríguez Iglesias, “FAIR approaches applied to unraveling plantpathogen interactions data and RNA processing evolution”, UPM, Spain
2013 Meifania Monica Chen, “Lipoprotein Ontology: A Formal Representation of Lipoproteins", Curtin University, Australia
2012 Jose Antonio Miñarro-Gimenez, “Entorno para la gestion semantica de informacion biomedica en investigacion traslacional". UM, Spain
2011 Doris Meja Avila, “Estrategia de interoperabilidad semantica en el contexto de integracion de conocimiento geografico y ambiental. Caso de aplicacion: Biodiversity Ontology". UPM, Spain
Supervisión estudiantes
2015 Salvador Alonso Martnez, “Imagen Docker para pipelines de Metagenomica", Bioinformatics MSc project. UM, Spain
2017 Denis Mishel Uchuari, prácticas externas, Grado en Ingeniería Informática de Gestión y Sistemas de Información, UPV/EHU
Divulgación
Software libre (GNU/linux) para biólogos. BioGaia 3, 2003 (Colegio Oficial de Biólogos de Euskadi).
¿Qué puede hacer la web semántica por la biología? BioGaia 7, 2009 (Colegio Oficial de Biólogos de Euskadi).
Meritos académicos
Estudios de postgrado
2009 Doctorado en informática, Universidad de Manchester, UK. Nota final: minor corrections
2004 Máster bioinformática, Universidad de Manchester, UK. Nota final: distinction
Estudios de grado
2003 Licenciatura biología UPV/EHU
2001 Estancia erasmus en Canterbury Christ Church University College, UK
Idiomas
Inglés
Nivel alto, hablado y escrito
Certificado IELTS 7,5 (== C2)
Uso continuo durante estancia erasmus, master, doctorado, postgrado Marie Curie, congresos, docencia, etc.
Euskara
Nivel alto, hablado y escrito
EGA, HABE 3 (== C1)
Programa docente
Minería de datos
Programa teórico
1.- Introducción
Panorama general de la minería de datos
Aplicaciones actuales
Clasificación de técnicas de análisis de datos
2.- Nociones teóricas básicas
Correlación y causalidad
Aprendizaje supervisado y no supervisado
“Underfitting vs overfitting”
“Bias vs variance”
“Curse of dimensionality”
3.- Aspectos metodológicos de un proceso típico de análitica de datos
Organización de un proyecto de manera rigurosa, documentada y reproducible
Fases: limpieza y filtrado, exploración, construcción de modelo, generación de resultados y optimización, validación
Aspectos prácticos a tener en cuenta
4.- Clasificadores basados en distancias: KNN
Funcionamiento básico
Ejemplos prácticos de aplicaciones
5.- Clasificación no supervisada: Clustering
Funcionamiento básico y algoritmos de clustering más comunes
Ejemplos prácticos de aplicaciones
6.- Redes bayesianas
Introducción a inferencia bayesiana
Aprendizaje de redes bayesianas
Ejemplos prácticos de aplicaciones
7.- Arboles de decisión y regresión
Introducción a arboles de decisión y regresión
Inducción de arboles, criterios de podado
Ejemplos prácticos de aplicaciones
8.- Redes Neuronales
Introducción a redes neuronales
Estructura, aprendizaje y adaptación
Capas y deep learning
Ejemplos prácticos de aplicaciones
9.- Support Vector Machines (SVMs)
Introducción teórica
Tipos de SVMs
Selección de modelo
Ejemplos prácticos de aplicaciones
10.- Técnicas de mezcla modelos
Bagging
Boosting
Random forests
11.- Reducción de dimensiones mediante PCA
Programa práctico
Prácticas por ordenador en parejas: analizar conjuntos de datos.
Resultados y explicación GitHub (https://github.com/).
Libreria scilit-learn (http://scikit-learn.org/stable/), plataforma Anaconda (https://www.continuum.io/anaconda-overview).
Metodología
|
Tipo de docencia |
M. (Magistral) |
GO (Prácticas ordenador)
|
|
Horas lectivas |
30 |
30 |
|
Horas de trabajo |
45 |
45 |
Sistema de evaluación
10% de la nota final: Concurso entre parejas de alumnos usando el sistema Kaggle (https://inclass.kaggle.com/).
40% de la nota final: Prácticas entregadas.
50% de la nota final: Exámen escrito.
Proyecto de investigación
Estrategias para publicar y consumir Grandes Datos sobre Metagenómica de acuerdo a los principios FAIR
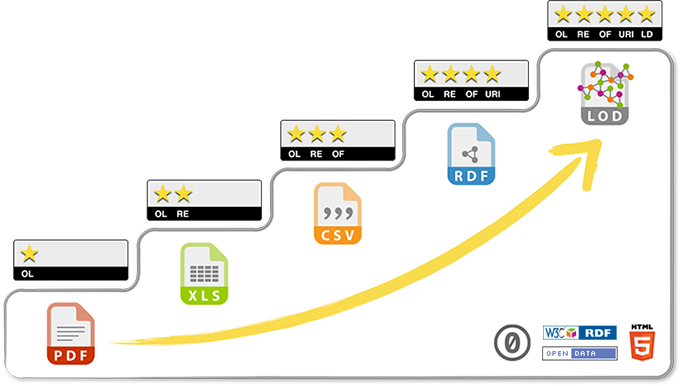
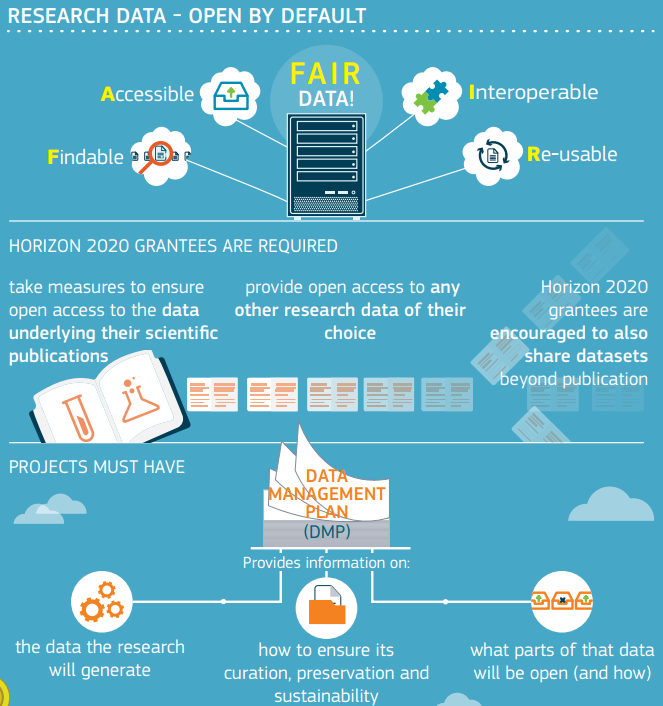
FAIR DATA
FINDABLE
→ Unambiguous identifiers supported by searchable metadata
ACCESSIBLE
→ Clearly-defined access protocol, preferably machine-actionable
INTEROPERABLE
→ Use shared vocabularies/ontologies in machine-accessible format
REUSABLE
→ Contextual information, allowing proper interpretation
→ Rich provenance information facilitating accurate citation
“Make adequate data stewardship mandatory
for all research proposals.
...Horizon 2020, should only support projects that properly address Data Stewardship... data infrastructures, that do not specify FAIR conditions...
should not be eligible for funding.”
Background

Linked Data
1.- Use URIs to identify entities and their relationships.
2.- Use HTTP URIs in order for agents to resolve (locate on the Web) such entities.
3.- When an entity is resolved via HTTP, information about the entity should be provided, in open standards. The format in which information is provided should be defined through content negotiation with the requesting agent (http://tools.ietf.org/html/rfc7231#section-5.3).
4.- The provided information should have HTTP resolvable typed links to other Linked Data resources, in order for the agent to discover more information by simply browsing through the links, as in “normal” Web browsing.
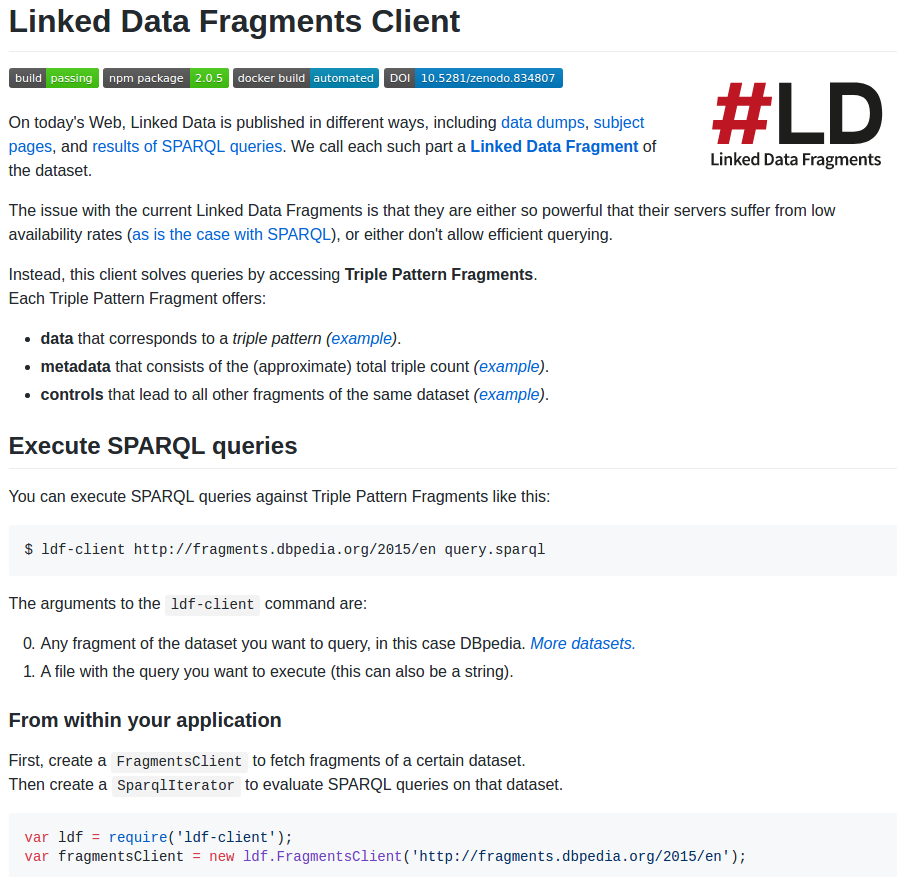
SADI

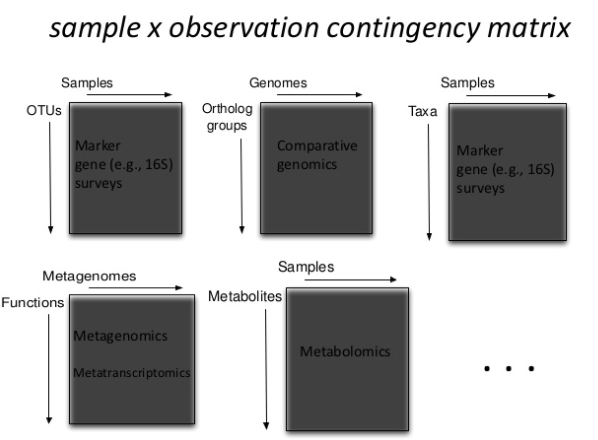
Big Data as Linked Data
1.- “Pure Linked Data” strategy
2.- “SADI Linked Data” strategy
3.- “Linked Data Fragments” strategy
1.- “Pure Linked Data” strategy

2.- “SADI Linked Data” strategy
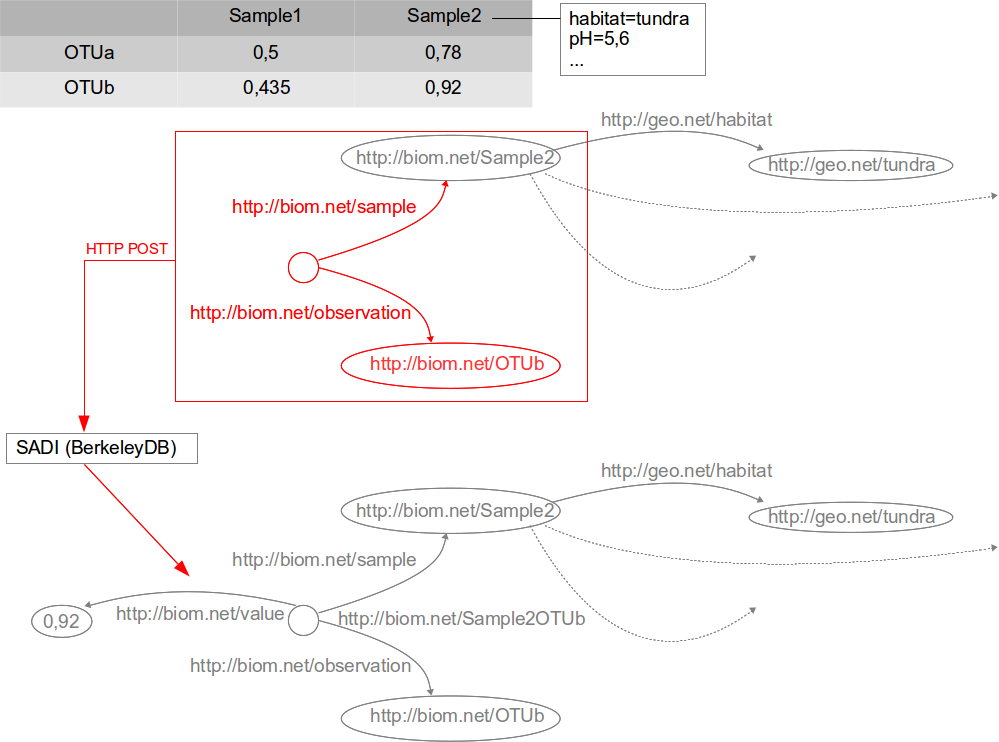
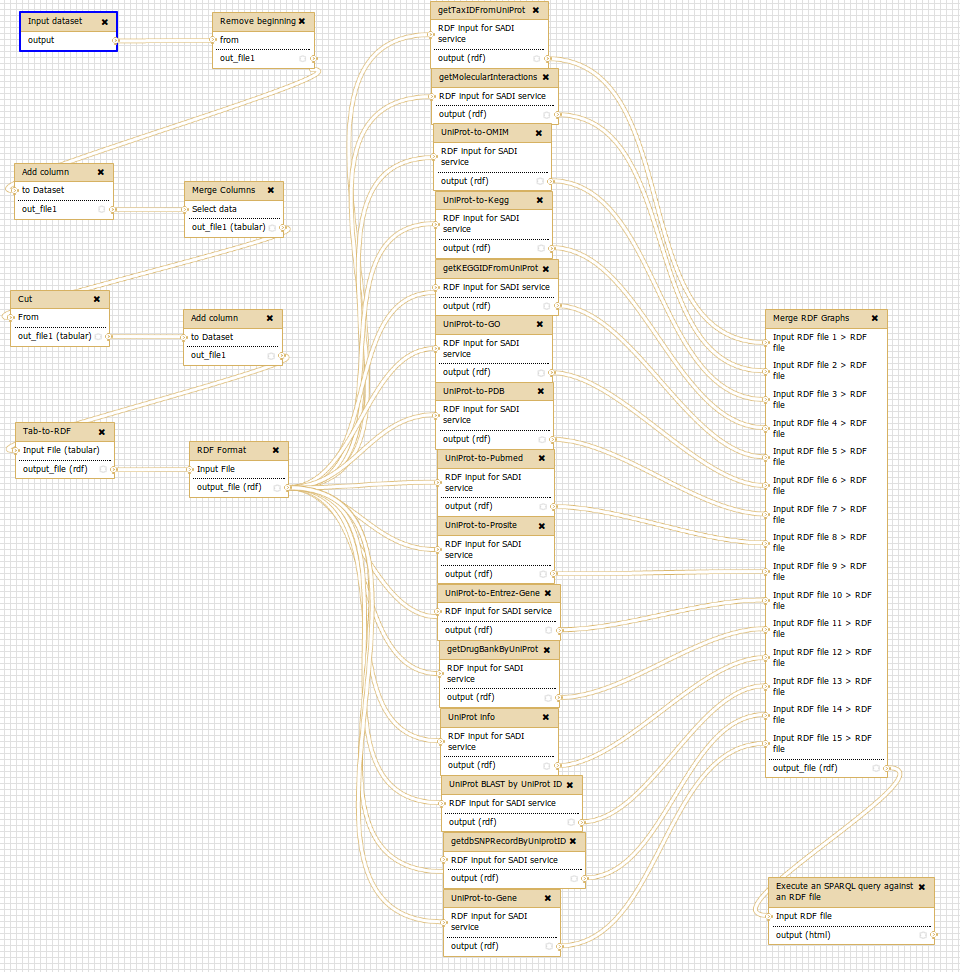

3.- “Linked Data Fragments” strategy
Big Data as Linked Data: specific targets
Develop HDF5 (BIOM) to RDF converter
Values (Key-Value Store / RDF )
Metadata (RDF)
Metadata links to LOD cloud (RDF)
Provenance
Convert BIOM to RDF
Publish RDF
Pure Linked Data
SADI Linked Data
Linked Data Fragments
Key-Value store
Triple Store
Link discovery
Evaluation
New queries due to integration
Performance