Approximate Bayesian Computation in Insurance
Patrick J. Laub and Pierre-Olivier Goffard


Background
- Undergrad (UQ): Software engineering & math
- PhD (Aarhus & UQ): Computational applied probability
- Post-doc #1 (ISFA): Insurance applications
- Post-doc #2 (UoM): Empirical dynamic modelling

A Software Engineer's Toolkit for Quant. Research

Motivation
Have a random number of claims \(N \sim p_N( \,\cdot\, ; \boldsymbol{\theta}_{\mathrm{freq}} )\) and the claim sizes \(U_1, \dots, U_N \sim f_U( \,\cdot\, ; \boldsymbol{\theta}_{\mathrm{sev}} )\).
We aggregate them somehow, like:
- aggregate claims: \(X = \sum_{i=1}^N U_i \)
- maximum claims: \(X = \max_{i=1}^N U_i \)
- stop-loss: \(X = ( \sum_{i=1}^N U_i - c )_+ \).
Question: Given a sample \(X_1, \dots, X_n\) of the summaries, what is the \(\boldsymbol{\theta} = (\boldsymbol{\theta}_{\mathrm{freq}}, \boldsymbol{\theta}_{\mathrm{sev}})\) which explains them?
E.g. a reinsurance contract
Likelihoods
For simple rv's we know their likelihood (normal, exponential, gamma, etc.).
When simple rv's are combined, the resulting thing rarely has a likelihood.
$$ X_1, X_2 \overset{\mathrm{i.i.d.}}{\sim} f_X(\,\cdot\,) \Rightarrow X_1 + X_2 \sim ~ \texttt{Unknown Likelihood}! $$
Have a sample of \(n\) i.i.d. observations. As \(n\) increases,
$$ p_{\boldsymbol{X}}(\boldsymbol{x} \mid \boldsymbol{\theta}) = \prod \text{Small things} \overset{\dagger}{=} 0, $$
or just takes a long time to compute, then \(\texttt{Intractable}\) \(\texttt{Likelihood}\)!
Usually it's still possible to simulate these things...
Approximate Bayesian Computation
Example: Flip a coin a few times and get \((x_1, x_2, x_3) = (\text{H, T, H})\); what is
Getting an exact match of the data is hard...
Accept fake data that's close to observed data
The 'approximate' part of ABC
Exact matching algorithm
Given some observations \(\boldsymbol{x}_{\text{obs}}\), repeat:
- generate a potential parameter from the prior distribution \(\boldsymbol{\theta}^{\ast} \sim \pi(\boldsymbol{\theta})\);
- simulate some 'fake data' \(\boldsymbol{x}^{\ast}\) from the model \(\boldsymbol{\theta}^{\ast}\);
- if \( \boldsymbol{x}_{\text{obs}} = \boldsymbol{x}^{\ast}\), then store \(\boldsymbol{\theta}^{\ast}\).
The resulting \(\boldsymbol{\theta}^{\ast}\)s are an i.i.d. sample from the posterior \(\pi(\theta \mid \boldsymbol{x}_{\text{obs}})\).
ABC acceptance–rejection
Given some observations \(\boldsymbol{x}_{\text{obs}}\), to generate \(K\) posterior samples:
- For \(k = 1\), \(\dots\), \(K\):
- Repeat:
- generate a potential parameter from the prior distribution \(\boldsymbol{\theta}^{\ast} \sim \pi(\boldsymbol{\theta})\);
- simulate some 'fake data' \(\boldsymbol{x}^{\ast}\) from the model \(\boldsymbol{\theta}^{\ast}\);
- Until \( \mathcal{D}( \boldsymbol{x}^{\ast} , \boldsymbol{x}_{\text{obs}} ) \leq \epsilon\)
- Store \(\boldsymbol{\theta}^{\ast}_k = \boldsymbol{\theta}^{\ast}\)
- Repeat:
Inputs: \(\mathcal{D}(\cdot, \cdot)\) is a distance measure, and the threshold \(\epsilon \ge 0\).
This samples from the approximate posterior a.k.a. the ABC posterior
$$ \pi_\epsilon(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) \propto \pi(\theta) \times \mathbb{P}\bigl( \mathcal{D}( \boldsymbol{x}^{\ast} , \boldsymbol{x}_{\text{obs}} ) \leq \epsilon \text{ where } \boldsymbol{x}^{\ast} \sim \boldsymbol{\theta} \bigr). $$Does it work in theory?
Rubio and Johansen (2013), A simple approach to maximum intractable likelihood estimation, Electronic Journal of Statistics.
Proposition: Say we have continuous data \( \boldsymbol{x}_{\text{obs}} \) , and our prior \( \pi(\boldsymbol{\theta}) \) has bounded support.
If for some \(\epsilon > 0\) we have
$$ \sup\limits_{ (\boldsymbol{z}, \boldsymbol{\theta}) : \mathcal{D}(\boldsymbol{z}, \boldsymbol{x}_{\text{obs}} ) < \epsilon, \boldsymbol{\theta} \in \boldsymbol{ \Theta } } \pi( \boldsymbol{z} \mid \boldsymbol{ \theta }) < \infty $$
then for each \( \boldsymbol{\theta} \in \boldsymbol{\Theta} \)
$$ \lim_{\epsilon \to 0} \pi_\epsilon(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) = \pi(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) . $$
We sample the approximate / ABC posterior
$$ \pi_\epsilon(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) \propto \pi(\theta) \times \mathbb{P}\bigl( \lVert \boldsymbol{x}_{\text{obs}}-\boldsymbol{x}^{\ast} \rVert \leq \epsilon \text{ where } \boldsymbol{x}^{\ast} \sim \boldsymbol{\theta} \bigr) , $$
but we care about the true posterior \( \pi(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) \) .
Mixed data
Get
$$ \lim_{\epsilon \to 0} \pi_\epsilon(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) = \pi(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) $$
when
We filled in some of the blanks for mixed data.
Our data was mostly continuous data but had an atom at 0.
Does it work in practice?
In other words, when does it break & how slow is it?
ABC Sequential Monte Carlo
- Input: data \(\boldsymbol{x}_{\text{obs}}\), prior \(\pi(\boldsymbol{\theta})\), distance \(\mathcal{D}(\cdot,\cdot)\)
# of generations \(G\), # of particles \(K\) - Start with \( \epsilon_0 = \infty \), \( \pi_0(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) = \pi(\boldsymbol{\theta}) \)
- For each generation \(g = 1\) to \(G\)
- For each particle \(k = 1\) to \(K\)
- Repeatedly:
- Generate a guess \( \boldsymbol{\theta}^{\ast} \) from \(\pi_{g-1}(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}})\)
- Generate fake data \(\boldsymbol{x}^{\ast}\) from \(\boldsymbol{\theta}^{\ast}\)
- Stop when \( \mathcal{D}(\boldsymbol{x}^\ast , \boldsymbol{x}_{\text{obs}}) < \epsilon_{g-1} \)
- Store \( \boldsymbol{\theta}_k^g = \boldsymbol{\theta}^{\ast} \)
- Repeatedly:
- Create a new threshold \(\epsilon_g \le \epsilon_{g-1} \) and a new population by discarding particles with \( \mathcal{D}(\boldsymbol{x}_k^g , \boldsymbol{x}_{\text{obs}}) \ge \epsilon_{g} \) until the effective sample size is \( K / 2 \)
- Weight each particle by \( w_k^g \propto \pi(\boldsymbol{\theta}_k^g) / \pi_{g-1}( \boldsymbol{\theta}_k^g \mid \boldsymbol{x}_{\text{obs}} ) \)
- Create a KDE \( \pi_g(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) \) based on the surviving \( ( \boldsymbol{\theta}_k^g , w_k^g ) \) particles
- For each particle \(k = 1\) to \(K\)
Dependent example
J. Garrido, C. Genest, and J. Schulz (2016), Generalized linear models for dependent frequency and severity of insurance claims, IME.
-
frequency \(N \sim \mathsf{Poisson}(\lambda=4)\),
-
severity \(U_i \mid N \sim \mathsf{DepExp}(\beta=2, \delta=0.2)\), defined as \( U_i \mid N \sim \mathsf{Exp}(\beta\times \mathrm{e}^{\delta N}), \)
-
summation summary \(X = \sum_{i=1}^N U_i \)
ABC posteriors based on 50 \(X\)'s and on 250 \(X\)'s given uniform priors.

ABC posterior vs true posterior
- Observe \(X = \sum_{i=1}^N U_i\) where \(N \sim \mathsf{Poisson}(\lambda=4)\), \(U_i \mid N \sim \mathsf{DepExp}(\beta=2, \delta=0.2)\)
- ABC posteriors (KDEs) based on 50 \(X\)'s and on 250 \(X\)'s.
- True posteriors (histograms) based on all the \(N\)'s and \(U_i\)'s used to make the 50 \(X\)'s and the 250 \(X\)'s.

Censored example
- frequency has negative binomial distribution \(N \sim \mathsf{NegBin}(\alpha=4, p=\frac23)\),
- severities have Weibull distributions \(U_i \sim \mathsf{Weibull}(k=\frac13, \beta=1)\),
- global stop-loss summary \(X = ( \sum_{i=1}^N U_i - 6 )_+\)
ABC posteriors based on 50 (11 non-zero) \(X\)'s and on 250 (69 non-zero) \(X\)'s with uniform priors.

Also observe frequency
Observe \(X\) and \(N\) where \(X = ( \sum_{i=1}^N U_i - 6 )_+\), \(N \sim \mathsf{NegBin}(\alpha=4, p=\frac23)\) and \(U_i \sim \mathsf{Weibull}(k=\frac13, \beta=1)\).
ABC posteriors based on 50 (11 non-zero) \(X\)'s and on 250 (69 non-zero) \(X\)'s with uniform priors.

Same figure zoomed in...

Misspecified example
- frequency has negative binomial distribution \(N \sim \mathsf{NegBin}(\alpha=4, p=\frac23)\),
- severities are fit with \(U_i \sim \mathsf{Gamma}(r, m)\) but generated by \(\mathsf{Weibull}(k=\frac13, \beta=1)\)
- global stop-loss summary \(X = ( \sum_{i=1}^N U_i - 6 )_+\)
ABC posteriors based on 50 (11 non-zero) \(X\)'s and on 250 (69 non-zero) \(X\)'s with uniform priors.

Model selection
Given some observations \(\boldsymbol{x}_{\text{obs}}\), to generate \(R\) samples from the posterior:
- For \(i = 1\), \(\dots\), \(R\):
- Repeat:
- choose a model \(m^{\ast} \sim \pi(m)\)
- generate a potential parameter from the prior distribution of that model \(\boldsymbol{\theta}^{\ast} \sim \pi(\boldsymbol{\theta} \mid m)\);
- simulate some 'fake data' \(\boldsymbol{x}^{\ast}\) from that parameter \(\boldsymbol{\theta}^{\ast}\);
- Until \( \lVert \boldsymbol{x}_{\text{obs}}-\boldsymbol{x}^{\ast} \rVert \leq \epsilon\)
- Store \(m^{\ast}_i = m^{\ast}\)
- Repeat:
Then the \(m^{\ast}\)s are samples from \(\pi(m \mid \boldsymbol{x}_{\text{obs}})\).
Model posterior: Weibull (✓) vs gamma (✗)?
ABC model posteriors based on 50 observations and on 250 observations.

Time-varying example
- Claims form a Poisson process point with arrival rate \( \lambda(t) = a+b[1+\sin(2\pi c t)] \).
- The observations are \( X_s = \sum_{i = N_{s-1}}^{N_{s}}U_i \).
- Frequencies are \(\mathsf{CPoisson}(a=1,b=5,c=\frac{1}{50})\) and sizes are \(U_i \sim \mathsf{Lognormal}(\mu=0, \sigma=0.5)\).



- Claims form a Poisson process point with arrival rate \( \lambda(t) = a+b[1+\sin(2\pi c t)] \).
- The observations are \( X_s = \sum_{i = N_{s-1}}^{N_{s}}U_i \).
- Frequencies are \(\mathsf{CPoisson}(a=1,b=5,c=\frac{1}{50})\) and sizes are \(U_i \sim \mathsf{Lognormal}(\mu=0, \sigma=0.5)\).
- ABC posteriors based on 50 \(X\)'s and on 250 \(X\)'s with uniform priors.
Wasserstein distance?
where \(\mathcal{S}_t\) denotes the set of all the permutations of \(\{1,\ldots, t\}\).
One-dimensional data:
Sorting is only \( \mathcal{O}(n \log n) \)

- Claims form a Poisson process point with arrival rate \( \lambda(t) = a+b[1+\sin(2\pi c t)] \).
- The observations are \( X_s = \sum_{i = N_{s-1}}^{N_{s}}U_i \).
- Frequencies are \(\mathsf{CPoisson}(a=1,b=5,c=\frac{1}{50})\) and sizes are \(U_i \sim \mathsf{Lognormal}(\mu=0, \sigma=0.5)\).
- ABC posteriors based on 50 \(X\)'s and on 250 \(X\)'s with uniform priors.

Compare \(\boldsymbol{x}_{\text{obs}} \sim f(\cdot, \boldsymbol{\theta}) \) to \(\boldsymbol{y} \sim f(\cdot, \boldsymbol{\theta}) \)

Normal Wasserstein distance ignores time

\( L^1 \) distance strictly enforces time
Balance \(t\) and \( x_t\)
\( x_1 \to ( 1, x_1) \)
\( x_2 \to ( 2, x_2) \)
The Wasserstein permutation is no longer just a sorting problem. To check exhaustively is \( \mathcal{O}(n!) \)
250! =
3232856260909107732320814552024368470994843717673780666747942427112823747555111209488817915371028199450928507353189432926730931712808990822791030279071281921676527240189264733218041186261006832925365133678939089569935713530175040513178760077247933065402339006164825552248819436572586057399222641254832982204849137721776650641276858807153128978777672951913990844377478702589172973255150283241787320658188482062478582659808848825548800000000000000000000000000000000000000000000000000000000000000
Traditional solution is the Hungarian algorithm \( \mathcal{O}(n^3) \)


Or drop back to univariate data using a Hilbert space-filling curve
https://blog.benjojo.co.uk/post/scan-ping-the-internet-hilbert-curve


https://xkcd.com/195/
- Claims form a Poisson process point with arrival rate \( \lambda(t) = a+b[1+\sin(2\pi c t)] \).
- The observations are \( X_s = \sum_{i = N_{s-1}}^{N_{s}}U_i \).
- Frequencies are \(\mathsf{CPoisson}(a=1,b=5,c=\frac{1}{50})\) and sizes are \(U_i \sim \mathsf{Lognormal}(\mu=0, \sigma=0.5)\).
- ABC posteriors based on 50 \(X\)'s and on 250 \(X\)'s with uniform priors.

We can scale time to balance the two axes
Bivariate example
- Two lines of business with dependence in the claim frequencies.
- Say \( \Lambda_i \sim \mathsf{Lognormal}(\mu \equiv 0, \sigma = 0.2) \).
- Claim frequencies \( N_i \sim \mathsf{Poisson}(\Lambda_i w_1) \) and \( M_i \sim \mathsf{Poisson}(\Lambda_i w_2) \) for \(w_1 = 15\), \(w_2 = 5\).
- Claim sizes for each line are \(\mathsf{Exp}(m_1 = 10) \) and \(\mathsf{Exp}(m_2 = 40)\).
- ABC posteriors based on 50 \(X\)'s and on 250 \(X\)'s with uniform priors.

Streftaris and Worton (2008), Efficient and accurate approximate Bayesian inference with an application to insurance data, Computational Statistics & Data Analysis
Time/cost
| Example | Time | Cost |
|---|---|---|
| Dependent | 45 s | 3.5 ¢ |
| Censored | 141 s | 11.1 ¢ |
| Misspecified | 40 s | 3.2 ¢ |
| Time-varying | ||
| Bivariate | ||
| TOTAL: |
|
|
On AWS c6g.16xlarge instance (64 ARM cores)
Architecture? Number of layers?
Number of neurons? Activation functions?
Skip-layer links? Normalisation layers?
Convolutional layers? Pre-processing input?
Test-train-validation split?

-
Input: data \(\boldsymbol{x}_{\text{obs}}\), prior \(\pi(\boldsymbol{\theta})\), distance \(\mathcal{D}(\cdot,\cdot)\)
# of generations \(G\), # of particles \(K\) - Start with \( \epsilon_0 = \infty \), \( \pi_0(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) = \pi(\boldsymbol{\theta}) \)
- For each generation \(g = 1\) to \(G\)
- For each particle \(k = 1\) to \(K\)
- Repeatedly:
- Generate a guess \( \boldsymbol{\theta}^{\ast} \) from \(\pi_{g-1}(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}})\)
- Generate fake data \(\boldsymbol{x}^{\ast}\) from \(\boldsymbol{\theta}^{\ast}\)
- Stop when \( \mathcal{D}(\boldsymbol{x}^\ast , \boldsymbol{x}_{\text{obs}}) < \epsilon_{g-1} \)
- Store \( \boldsymbol{\theta}_k^g = \boldsymbol{\theta}^{\ast} \)
- Repeatedly:
- Create a new threshold \(\epsilon_g \le \epsilon_{g-1} \) and a new population by discarding particles with \( \mathcal{D}(\boldsymbol{x}_k^g , \boldsymbol{x}_{\text{obs}}) \ge \epsilon_{g} \) until the effective sample size is \( K / 2 \)
- Weight each particle by \( w_k^g \propto \pi(\boldsymbol{\theta}_k^g) / \pi_{g-1}( \boldsymbol{\theta}_k^g \mid \boldsymbol{x}_{\text{obs}} ) \)
- Create a KDE \( \pi_g(\boldsymbol{\theta} \mid \boldsymbol{x}_{\text{obs}}) \) based on the surviving \( ( \boldsymbol{\theta}_k^g , w_k^g ) \) particles
- For each particle \(k = 1\) to \(K\)
- Return the final population \(\{(\boldsymbol{\theta}_k^G , w_k^G)\}_{k=1,\dots,K}\)
Transform (e.g. take logarithms)?
Which distributions?
What distance?
Systematic resampling?
Generate just one fake data?
Accept fake data with some probability?
Fixed population size? (Keep a quantile)
Adjust final population using linear regression?
\(w_{\cdot}^{g-1}\) ?
Summarise data?
ABC turns a statistics problem into a programming problem
... to be used as a last resort
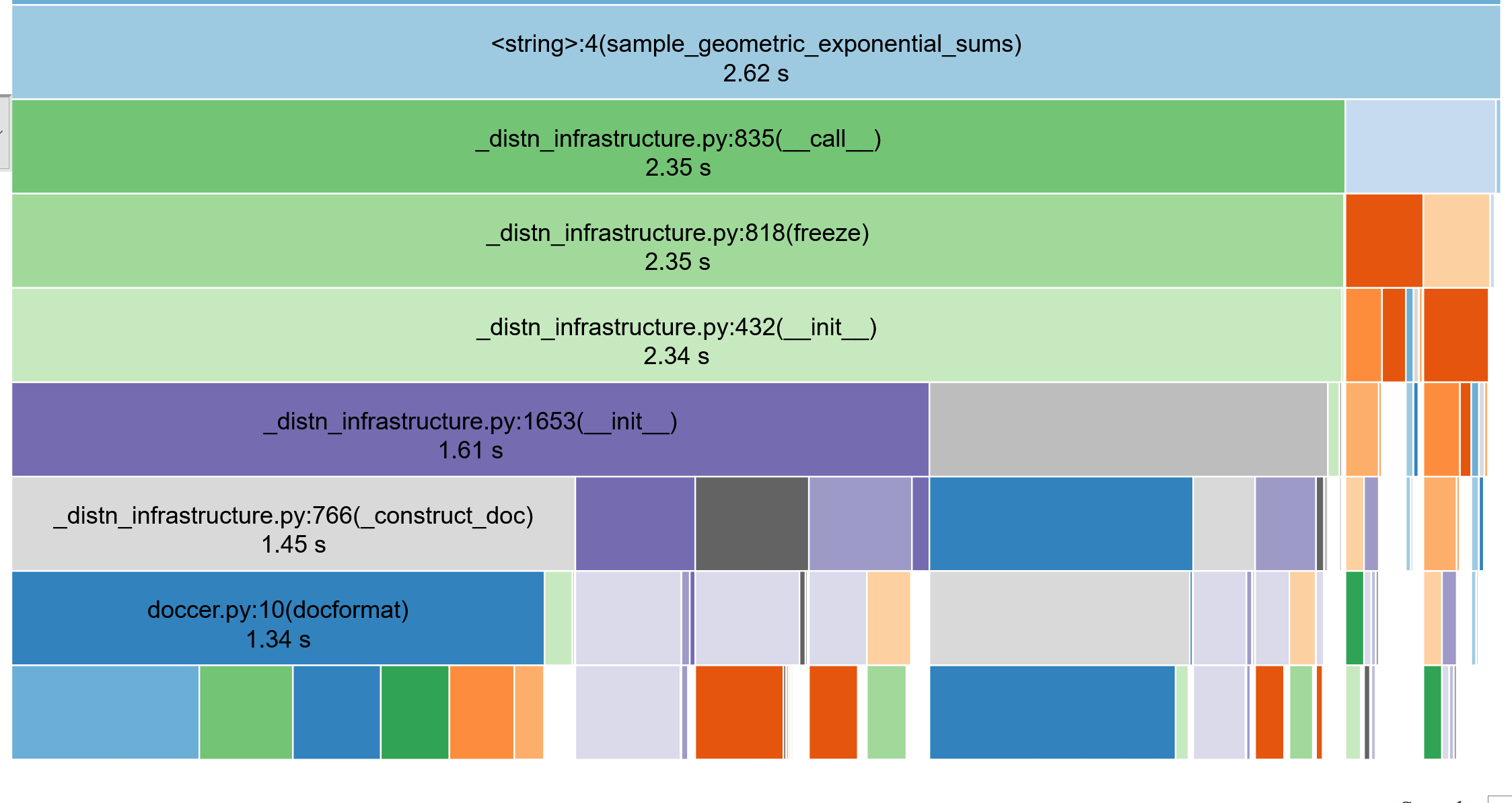
Profiling the code
(use snakeviz, lineprofiler)
from scipy import stats
def sample_geometric_exponential_sums(T, p, μ):
result = []
for t in range(T):
N_t = stats.geom(p).rvs()
X_t = 0.0
for n in range(N_t):
U_n = stats.expon(μ).rvs()
X_t += U_n
result.append(X_t)
return resultTypically need to call this \(10^7\) times
N_t = stats.exp(mu).rvs()N_t = stats.exp.rvs(mu)1.7 s vs 89 ms ... (19x speedup)

import numpy.random as rnd
def sample_geometric_exponential_sums(T, p, μ):
result = []
for t in range(T):
N = rnd.geometric(p)
X_t = 0.0
for n in range(N):
U_n = rnd.exponential(μ)
X_t += U_n
result.append(X_t)
return resultLooking in the source, see that scipy just wraps numpy
Down from 89 ms to 5.5 ms (another 16x speedup)
Vectorised code
(use numpy)
Benefits of vectorised code
- Readability; more concise, expressing high-level ideas, e.g.
- Replace slow interpreted loops with fast compiled code
- Potentially can use a CPU's vectorised instructions (SIMD)
- Better use of the CPU's cache
\[ \sum_{n=0}^{\infty} \frac{f^{(n)}(a)}{n!} (x-a)^n \]
def sample_geometric_exponential_sums(T, p, μ):
result = []
for t in range(T):
N = rnd.geometric(p)
X_t = 0.0
for n in range(N):
X_t += rnd.exponential(μ)
result.append(X_t)
return resultUnvectorised
def sample_geometric_exponential_sums(T, p, μ):
X = np.zeros(T)
N = rnd.geometric(p, size=T)
for t in range(T):
X[t] = rnd.exponential(μ, size=N[t]).sum()
return XVectorised (& most readable)
5.5 ms to 5.48 ms (no change)
def sample_geometric_exponential_sums(T, p, μ):
X = np.zeros(T)
N = rnd.geometric(p, size=T)
U = rnd.exponential(μ, size=N.sum())
i = 0
for t in range(T):
X[t] = U[i:i+N[t]].sum()
i += N[t]
return X5.5 ms to 2.7 ms (2x speedup)
Allocate all random variables in one hit
Compiled code
(use numba)
Interpreters & compilers
C.f. Lin Clark (2017), A crash course in JIT compilers

Why is Python slow?
x = 1
y = 2
z = x + yint x = 1;
int y = 2;
int z = x + y;// x = 1
if (PyDict_SetItem(__pyx_d, __pyx_n_s_x, __pyx_int_1) < 0) {
__PYX_ERR(0, 2, __pyx_L1_error);
}
// y = 2
if (PyDict_SetItem(__pyx_d, __pyx_n_s_y, __pyx_int_2) < 0) {
__PYX_ERR(0, 3, __pyx_L1_error);
}
// z = x + y
__Pyx_GetModuleGlobalName(__pyx_t_1, __pyx_n_s_x);
if (unlikely(!__pyx_t_1)) {
__PYX_ERR(0, 4, __pyx_L1_error);
}
__Pyx_GOTREF(__pyx_t_1);
__Pyx_GetModuleGlobalName(__pyx_t_2, __pyx_n_s_y);
if (unlikely(!__pyx_t_2)) {
__PYX_ERR(0, 4, __pyx_L1_error);
}
__Pyx_GOTREF(__pyx_t_2);
__pyx_t_3 = PyNumber_Add(__pyx_t_1, __pyx_t_2);
if (unlikely(!__pyx_t_3)) {
__PYX_ERR(0, 4, __pyx_L1_error);
}
__Pyx_GOTREF(__pyx_t_3);
__Pyx_DECREF(__pyx_t_1);
__pyx_t_1 = 0;
__Pyx_DECREF(__pyx_t_2);
__pyx_t_2 = 0;
if (PyDict_SetItem(__pyx_d, __pyx_n_s_z, __pyx_t_3) < 0) {
__PYX_ERR(0, 4, __pyx_L1_error);
}
__Pyx_DECREF(__pyx_t_3);
__pyx_t_3 = 0;Python
C++
def arraySum(arr):
sum = 0;
for i in range(len(arr)):
sum += arr[i];
return sum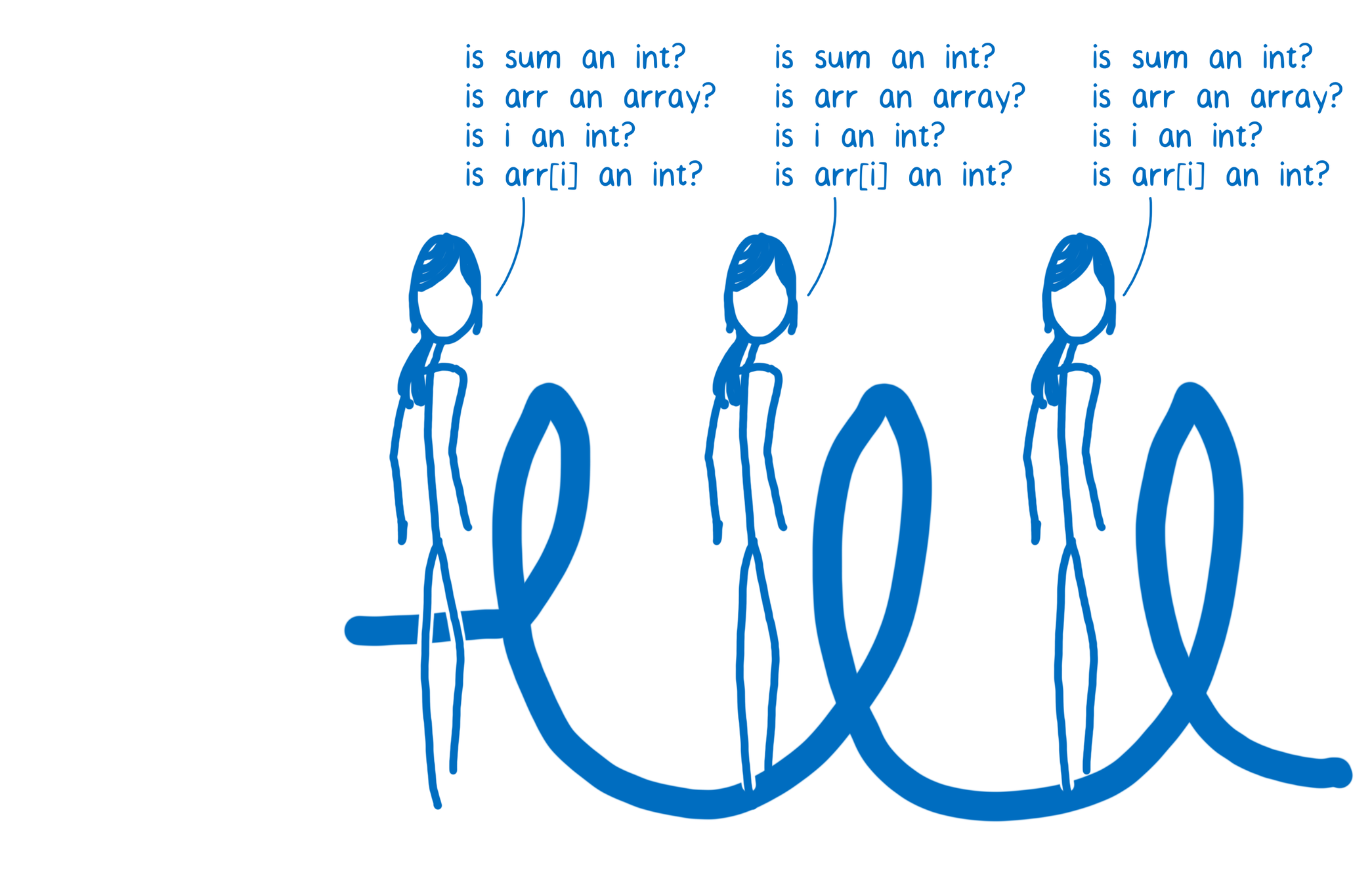
C.f. Lin Clark (2017), A crash course in JIT compilers
https://hacks.mozilla.org/2017/02/a-crash-course-in-just-in-time-jit-compilers/
def sample_geometric_exponential_sums(T, p, μ):
X = np.zeros(T)
N = rnd.geometric(p, size=T)
U = rnd.exponential(μ, size=N.sum())
i = 0
for t in range(T):
X[t] = U[i:i+N[t]].sum()
i += N[t]
return XInterpreted version
from numba import njit
@njit()
def sample_geometric_exponential_sums(T, p, μ):
X = np.zeros(T)
N = rnd.geometric(p, size=T)
U = rnd.exponential(μ, size=N.sum())
i = 0
for t in range(T):
X[t] = U[i:i+N[t]].sum()
i += N[t]
return XFirst run is compiling (500 ms), but after we are
down from 2.7 ms to 164 μs (16x speedup)
Compiled by numba
Original code: 1.7 s
Basic profiling with snakeviz: 5.5 ms, 310x speedup
+ Vectorisation/preallocation with numpy: 2.7 ms, 630x speedup
+ Compilation with numba: 164 μs, 10,000x speedup
And potentially:
+ Parallel over 80 cores: say another 50x improvement,
so overall 50,000x speedup.
Parallel code
(use joblib)
Computing in parallel
serialResult = sum(f(i) for i in range(n+1))
from joblib import Parallel, delayed
with Parallel(n_jobs=10) as parallel:
parResult = sum(parallel(delayed(f)(i) for i in range(n)))In python, use joblib. E.g. to calculate
\[ \sum_{i=0}^{n-1} f(i) \]
In practice..
Either:
- too much parallelism for the computer
- a problem which is too small for the parallelism
Number of cores
Execution Time
Contributions
- Modest theoretical contribution, looking at convergence of the approximate posterior to the true posterior in the case of mixed data (e.g. truncated data).
- An optimized parallel Python package abcre ('re' for reinsurance), on Github
Take-home messages
- What is ABC? What can it do?
- When should it be used?
- How does it fail?
- ABC turns a statistics problem into a programming problem
- Profile (snakeviz), vectorise (numpy), compile (numba), and parallelise your code (joblib)
Appendix
Bayesian statistics
Prior distribution \(\pi(\boldsymbol{\theta}) \)
Likelihood \(\pi( \boldsymbol{x} \mid \boldsymbol{\theta} )\)
Posterior distibution
$$ \pi(\boldsymbol{\theta} \mid \boldsymbol{x} ) = \frac{ \pi(\boldsymbol{\theta}) \pi( \boldsymbol{x} \mid \boldsymbol{\theta} ) }{ \pi( \boldsymbol{x} ) } $$

Simulation results
But first, what does a success look like?
Markov chain Monte Carlo


C.f. Lin Clark (2017), A crash course in JIT compilers
https://hacks.mozilla.org/2017/02/a-crash-course-in-just-in-time-jit-compilers/
Talking to a computer
def sample_geometric_exponential_sums(T, p, μ):
X = np.zeros(T)
N = rnd.geometric(p, size=T)
U = rnd.exponential(μ, size=N.sum())
i = 0
for t in range(T):
X[t] = U[i:i+N[t]].sum()
i += N[t]
return XInterpreted version
cpdef sample_geometric_exponential_sums(long T, double p, double mu):
cdef double[:] X = np.zeros(T)
cdef long[:] N = rnd.geometric(p, size=T)
cdef double[:] U = rnd.exponential(mu, size=np.sum(N))
cdef int i = 0
cdef int t
for t in range(T):
X[t] = np.sum(U[i:i+N[t]])
i += N[t]
return XCompiled by Cython (not recommended)
Writing faster code
"Premature optimization is the root of all evil." Donald Knuth
Stop and ask why
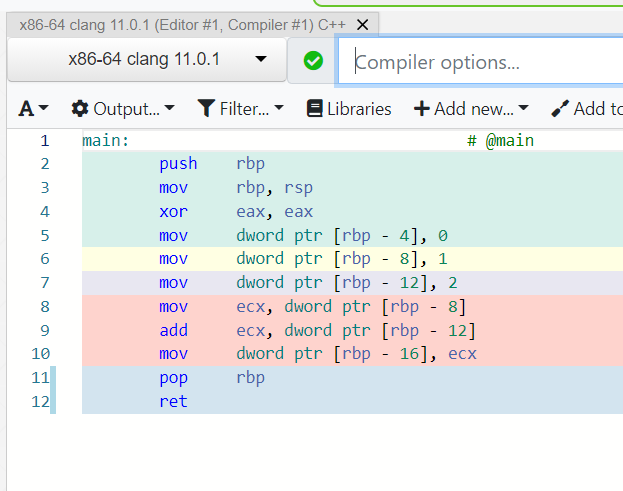
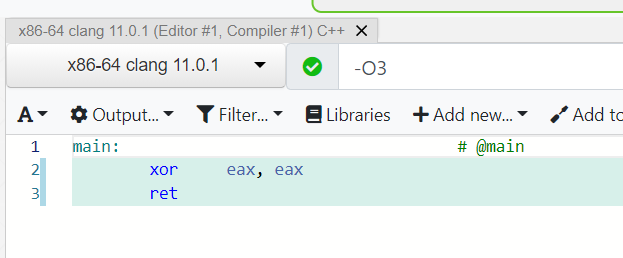
What does a machine understand?
Intel i5 (2010): add (0101), subtract (1111), ...
Intel i7 (2020): add (0101), subtract (1111), add_vectors (1011), ...
Apple M1 (2020): add (100), subtract (111), ...
movl $0x1, -0x8(%rbp)int x = 1;Instruction set architecture (ISA), e.g. x86-64
Try Compiler Explorer https://godbolt.org/z/dTv78M
A typical computer



A typical program
Time ->
Waiting to read or write data to memory

Source: Joel Fenwick's CSSE2310 lecture slides
Running


Multiple threads or multiple processes?
Use multiple processes in Python,
probably should use multiple threads elsewhere.

Baumann et al. (2019), A fork() in the road
17th Workshop on Hot Topics in Operating Systems


Amdahl’s Law (or Law of Diminishing Returns)
\(P\) is the proportion of a program that can be made parallel
\( 1 - P \) remains serial.
Theoretical maximum speedup of achieved by N processor is
\[ S(N)=1/[(1-P)+P/N] \]
Hence with unlimited processor count
\[ S(\infty) = 1 / (1-P) \]
E.g. \(P = 0.9\) then \( S(\infty) = 10 \).
Theoretical limits
Know your CPU

Intel lies
Apple confuses

>>> import psutil
>>> psutil.cpu_count()
4
>>> psutil.cpu_count(logical=False)
2Apple is cooler (literally)
# This tells Python to just use one thread for internal
# math operations. When it tries to be smart and do multithreading
# it usually just stuffs everything up and slows us down.
import os
for env in ["MKL_NUM_THREADS", "NUMEXPR_NUM_THREADS", "OMP_NUM_THREADS"]:
os.environ[env] = "1"
sample_abc_population(N)
sample_one()
sample_one()
sample_one()
sample_one()
np.solve(A, x)

parfor i = 1:m
for j = 1:n
f(i, j)
end
endfor i = 1:m
parfor j = 1:n
f(i, j)
end
end

11.9 s
41.8 s
class Parallel(Logger):
"""
...
batch_size: int or 'auto', default: 'auto'
The number of atomic tasks to dispatch at once to each
worker. When individual evaluations are very fast, dispatching
calls to workers can be slower than sequential computation because
of the overhead. Batching fast computations together can mitigate
this.
The ``'auto'`` strategy keeps track of the time it takes for a batch
to complete, and dynamically adjusts the batch size to keep the time
on the order of half a second, using a heuristic. The initial batch
size is 1.
``batch_size="auto"`` with ``backend="threading"`` will dispatch
batches of a single task at a time as the threading backend has
very little overhead and using larger batch size has not proved to
bring any gain in that case.
...
"""https://github.com/joblib/joblib/blob/master/joblib/parallel.py