Accelerating Edge Metagenomic Analysis with Serverless-based Cloud Offloading
Piotr Grzesik
dr hab. inż. Dariusz Mrozek, prof. PŚ
Silesian University of Technology
Agenda
- Metagenomic and Nanopore Sequencing
- MinION Nanopore
- Challenges of metagenomics at the edge
- Edge-based approach
- Serverless computing
- Serverless approach
- Hybrid approach
- Summary
- Next steps
Metagenomics analysis
Metagenomics - study of genetic material from environmental samples. It is used to sequence DNA (or RNA) based on the prepared sample, identify microorganisms, detect potential mutations or identify previously unknown species.
Nanopore sequencing
Nanopore sequencing - developed by Oxford Nanopore Technologies, it is a process of DNA sequencing that works by monitoring changes to an electrical current caused by DNA strand passing through a nanopore. The signal that is obtained as a result is decoded to specific DNA or RNA sequences. The process of such decoding is called basecalling.
Nanopore sequencing

MinION Nanopore
MinION Nanopore - portable sequencing device, released by Oxford Nanopore Technologies in 2014. It is the first device that enables portable sequencing at affordable price (1000$). It is powered via USB, weights under 100g, which makes it possible to use it as a field device.
MinION Nanopore

Metagenomics at the edge
Edge computing is a computing paradigm that brings the data processing and storage closer to a place where it is needed. It allows to reduce the volume of data that needs to be send over the Internet, allows to improve reaction time to the changing state of the system and improves resilience and allows for data loss prevention where Internet connection is not reliable or not available at all most of the time.
Challenges of metagenomics at the edge
- Computational complexity
- Volume of data that needs to be processed
- Limited network connectivity
- Limited access to power supplies
Analysis workflow
The considered analysis workflow consist of three separate steps - sequencing, basecalling and classification. During the first step, MinION device will sequence DNA, outputting electrical current measurements in form of FAST5 files. In the second step, basecaller will interpret electrical current and output genomic sequences. In the last step, genomic sequences will be classified and labeled with taxonomic labels.

Testing environment

Testing environment
-
Simulated MinION Nanopore that outputs reads in form of fast5 files
-
Edge device that runs basecalling and classification analysis with SSD Disk
-
Cloud-based service, long-term storage of sequenced data
-
Power supply with INA219 + Raspberry Pi 4 for current measurement
Edge device specification
- Single board computer - Jetson Xavier NX
-
GPU - NVIDIA Volta™ architecture
with 384 NVIDIA® CUDA® cores and 48 Tensor cores -
6-core NVIDIA Carmel ARM®v8.2 64-bit CPU
6 MB L2 + 4 MB L3 - 8 GB 128-bit LPDDR4x 51.2GB/s
- Storage - SDHC card (32 GB, class 10) + SSD NVMe
- OS - Ubuntu 18.04.5 LTS with kernel version 4.19.140-tegra
Edge device specification

Software used
- Guppy - It is state-of-the-art, closed-source, basecaller developed by Oxford Nanopore Technologies, creator of MinION Nanopore device. It offers support for both CPU and GPU acceleration on selected devices, works both on AArch64 and x86-64 architectures. It also supports two different modes - fast and hac, which stands for high accuracy.
- Kraken 2 - taxonomic classification system using exact k-mer matches to achieve high accuracy and fast classification speeds. This classifier matches each k-mer within a query sequence to the lowest common ancestor (LCA) of all genomes containing the given k-mer. Due to significant RAM use of standard Kraken databases, a new, minimized database was prepared, dedicated for identification of bacterias and virals.
Experiments
-
Fast5 files with data from sequencing runs containing material of Escherichia coli and Klebsiella Pneumoniae
-
Measurement of samples processed per second in different power modes
-
Guppy and Bonito was tested with and and without GPU acceleration enabled
-
During each run, current was measured with INA219 + Raspberry Pi 4 circuit
-
Classification with Kraken2 was also ran in different power modes
-
Available power modes in Jetson Xavier NX: 15W 6 CORE, 15W 4 CORE, 15W 2 CORE, 10W 2 CORE, 10W 4 CORE
Results

Samples per second processed by Guppy with Fast model
Results
Samples per second processed by Guppy with HAC model

Results
Average power for Guppy with Fast model

Results
Average power for Guppy with HAC model

Observations
-
Guppy with Fast model is able to support real-time basecalling for up to 3 MinION devices at the same time
-
Jetson Xavier NX is a sutiable device for running such experiments in places with limited network connectivity and limited power supplies
-
Using 10W power mode on Jetson Xavier NX is more efficient energy-wise than 15W power mode
-
CPU basecalling is not feasible on Jetson Xavier NX device (on similar ones as well)
-
As of right now, it seems like Guppy is the only basecaller suitable to use at the edge
Serverless computing
Serverless computing is a computing paradigm that takes advantage of simple, stateless functions (also called Functions-as-a-service) that offer low maintenance overhead, fault tolerance, support massive parallelism, allocate resources on-demand and can quickly scale both up and down. One additional benefit of this paradigm is that users pay only for actual invocations of functions and not for idle time.
Serverless computing
Serverless computing is also getting more popular in the literature for bioinformatic purposes:
- sVEP (Serverless Variant Effect Predictor) - Cloud Native Variant Annotation Pipeline - speedup vs traditional VM-based solutions
- GT-Scan2 - Serverless Target Finder for genome editing - more cost-effective than traditional VM-based solutions
- Serverless all-against-all Pairwise comparison using Smith-Waterman algorithm - both faster and more cost-effective than traditional VM-based solutions
AWS Lambda
- Serverless computing platform, introduced in 2014 by Amazon Web Services
- Out of the box support for Java, .NET Core, Go, Ruby, Python, Node.js. Virtually supports every runtime via custom runtimes introduced in 2018
- Easy integration with S3, Kinesis, CloudWatch, API Gateway
- Execution time limit - 15 minutes
- Alternatives: Google Cloud Functions, Microsoft Azure Functions, Apache OpenWhisk
Why Lambda for bioinformatics?
(Added at the end of 2020)
- Support for up to 10,240 MBs of memory
- Support for up to 6 virtual CPU cores
- Support for Docker containers
- Support for packages with up to 10 GB in size
- Support for AVX2 instruction set
Analysis workflow

Analysis workflow
In proposed workflow, first step is uploading FAST5 files from MinION Nanopore to S3 Bucket. In the next step, the processing is triggered manually and first Lambda function splits the FAST5 files into batches and schedules execution of multiple Lambda functions that run basecalling operation and save results to S3 bucket as well.
Software used
- Guppy - It is state-of-the-art, closed-source, basecaller developed by Oxford Nanopore Technologies, creator of MinION Nanopore device. It offers support for both CPU and GPU acceleration on selected devices, works both on AArch64 and x86-64 architectures. It also supports two different modes - fast and hac, which stands for high accuracy.
Experiments
-
Fast5 files with data from sequencing runs containing material of Escherichia coli and Klebsiella Pneumoniae
-
Measurement of samples processed per second and per second per MB of memory for different models
-
Both Guppy and Bonito were tested
-
Experiments were run for 256, 512, 1024, 2048, 4096, 6144, 8192 and 10240 (maximum) MBs of RAM available to a single Lambda function
Results
Samples per second processed by Guppy with Fast model

Results
Samples per second per MB of memory for Guppy fast model

Results
Samples per second processed by Guppy with HAC model

Results
Samples per second per MB of memory for Guppy high accuracy model

Observations
- Guppy CPU basecaller is offering the best performance
- Other basecallers are either very slow (Bonito) or require significant adjustments (Deepnano-blitz)
- Takes advantage of recently introduced container support in Lambda
- Scaled 100 simultaneous functions, each with 6 vCPUs in < 1 minute
- At most each function is able to process ~600000 samples/s on average, which given theoretical maximum output of MinION (2,300,000 signals/s) allows 3-4 functions to basecall data in near real-time
Hybrid approach
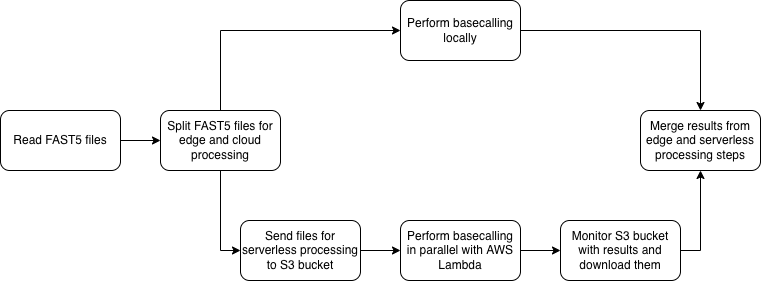
Hybrid approach
In proposed workflow, first step is to determine if we can take advantage of cloud offloading to speed up the edge processing. Then, the files are splitted into batches for edge and serverless processing, based on the theoretical processing speeds for both approaches, depending on upload speed. Then, the files are processed separately, and edge device monitors and collects the results from cloud-based processing as well.
Experiments
-
Fast5 files with data from sequencing runs containing material of Escherichia coli and Klebsiella Pneumoniae
-
Jetson Xavier NX as edge device, tested with lowest (10W 2 core) and highest (15W 6 core) power modes
-
Guppy basecaller was tested
-
Experiments were run for 128, 256, 512 kB/s upload speeds
Results
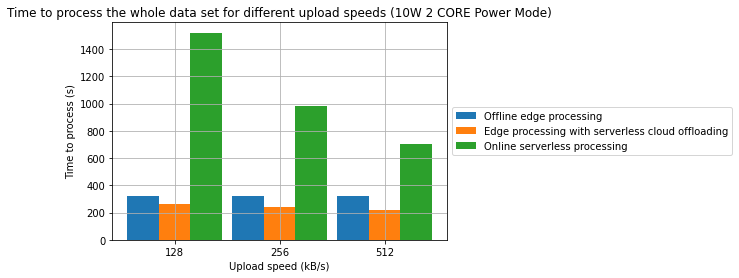
Results
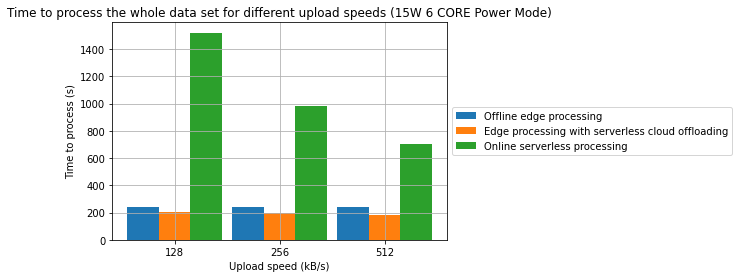
Results
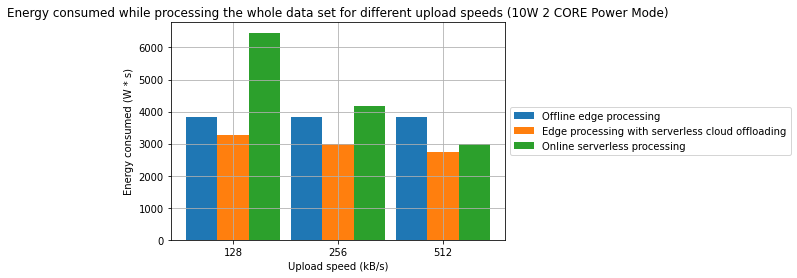
Results
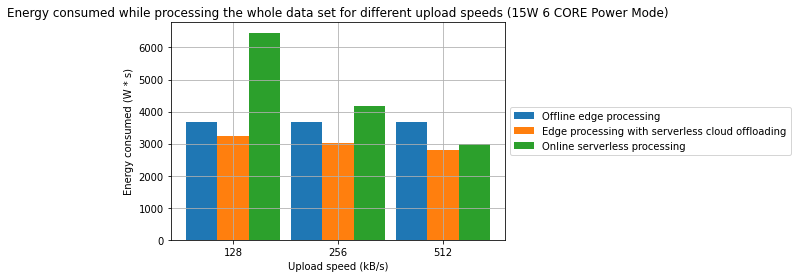
Observations
- A combination of offline edge processing with online serverless processing can be an effective strategy for reducing energy consumption and processing time
- In one of the cases, we have managed to achieve a 31.2% reduction in processing time and in the other 28.7% reduction in energy consumption in comparison to a fully offline process
- From an energy standpoint, in some cases, fully online processing might be more energy effective than offline processing