GeSubNet: GENE INTERACTION INFERENCE FOR DISEASE SUBTYPE NETWORK GENERATION
VAE Models for lower dim... embed... and survival pre...
Subhankar Mishra, Sachin Kumar
BL2403, School of Computer Sciences, NISER


GeSubNet: GENE INTERACTION INFERENCE FOR DISEASE SUBTYPE NETWORK GENERATION
Introduction
This paper studies the design of an architecture to predict cancer type.
/* Big handwritten title */
h1, h2 {
font-family: 'Permanent Marker', cursive;
font-weight: bold;
}
/* Orange banner for section titles */
.section-title {
background-color: orange;
color: black;
padding: 10px;
display: inline-block;
}
/* Author text style */
.authors {
color: darkred;
font-family: 'Shadows Into Light', cursive;
font-size: 1.4em;
}
/* Footer/affiliation */
.affiliation {
font-family: monospace;
font-size: 1.1em;
}
There are two knowledge databases
Problem
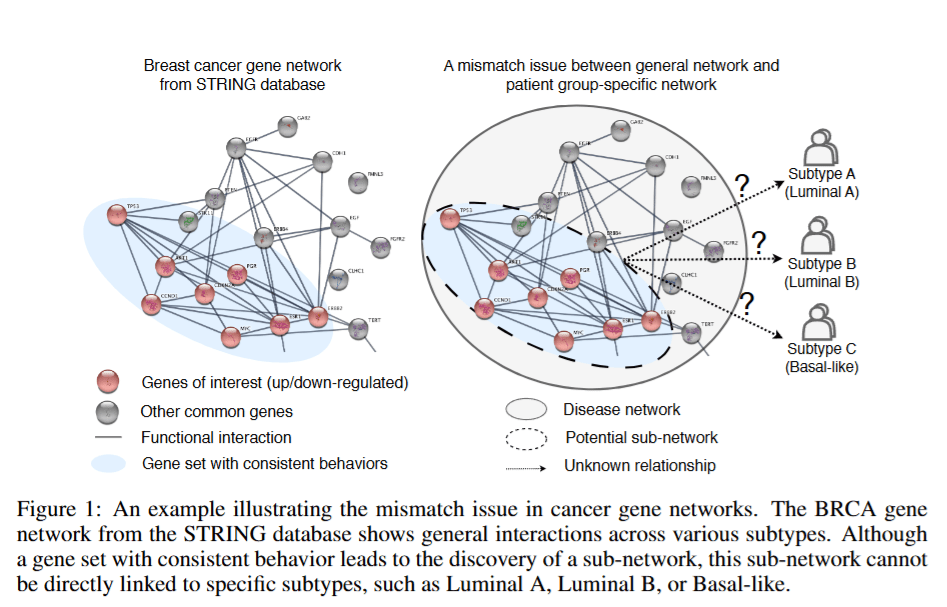
Related Work
Two main approaches exist for building subtype gene networks: statistical and deep learning-based.
Statistical methods use correlation metrics (e.g., Pearson, mutual information) to detect co-expressed or functionally linked genes.
Deep learning methods build gene networks using graph neural networks (GNNs), embedding patient data and predicting gene interactions.
Most deep learning models focus on general disease associations, failing to capture subtype-specific gene interactions.
PRELIMINARY AND PROBLEM SETTING
Problem setting
\textbf{Definition 1 (Gene expression data).} The fundamental entity in gene expression profile data is the individual patient. Each patient profile comprises tens of thousands of genes with measured features.
Let
\[
X = \{x^{(m)}\}_{m=1}^M
\]
denote a dataset of $M$ patients. Each patient can be represented as a sequence of $N$ gene measurements:
\[
x^{(m)} = \{x^{(m)}_1, x^{(m)}_2, \dots, x^{(m)}_N\}.
\]
Let
\[
Y = \{y_1, y_2, \dots, y_{|Y|}\}
\]
denote the set of subtypes for a cancer. Each $x^{(m)}$ is associated with a label $y$.